

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148484.5 - phase: 0
(519 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
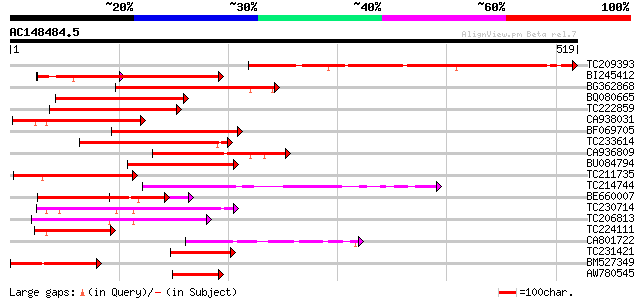
Score E
Sequences producing significant alignments: (bits) Value
TC209393 similar to UP|Q9FYW4 (Q9FYW4) BAC19.12, partial (26%) 332 2e-91
BI245412 homologue to GP|29028906|gb At2g02080 {Arabidopsis thal... 257 8e-69
BG362868 homologue to PIR|T46147|T461 zinc finger protein - Arab... 250 1e-66
BQ080665 238 4e-63
TC222859 homologue to GB|AAO64832.1|29028906|BT005897 At2g02080 ... 228 6e-60
CA938031 201 5e-52
BF069705 similar to GP|16226322|gb AT3g45260/F18N11_20 {Arabidop... 200 1e-51
TC233614 190 1e-48
CA936809 homologue to PIR|T46147|T461 zinc finger protein - Arab... 184 1e-46
BU084794 182 3e-46
TC211735 178 6e-45
TC214744 homologue to UP|Q9LVQ7 (Q9LVQ7) Zinc finger protein, pa... 167 8e-42
BE660007 similar to GP|29028906|gb At2g02080 {Arabidopsis thalia... 147 9e-36
TC230714 weakly similar to GB|AAO44076.1|28466935|BT004810 At5g2... 109 3e-24
TC206813 similar to UP|CW14_YEAST (O13547) Covalently-linked cel... 106 3e-23
TC224111 similar to UP|Q944L3 (Q944L3) AT3g45260/F18N11_20 (ID1-... 105 4e-23
CA801722 102 5e-22
TC231421 homologue to GB|AAM10292.1|20147151|AY091693 AT3g13810/... 99 5e-21
BM527349 90 3e-18
AW780545 89 4e-18
>TC209393 similar to UP|Q9FYW4 (Q9FYW4) BAC19.12, partial (26%)
Length = 1422
Score = 332 bits (852), Expect = 2e-91
Identities = 207/312 (66%), Positives = 231/312 (73%), Gaps = 11/312 (3%)
Frame = +2
Query: 219 PAVITTTAAAAATTPQSNSGVSSALETQKLDLPE-NPPQIIEE-PQVVVTTTASALNASC 276
P + +A A TT Q+NS +S L+TQ L+LPE NPPQ+IEE PQ A+A++ SC
Sbjct: 2 PPLQPVSATVATTTTQTNSAMSCGLQTQNLELPETNPPQVIEEEPQ-----GATAVSGSC 166
Query: 277 -SSSTSSKSNGCAAT----STGVFASLFASSTASASASLQPQAPAFSDLIRSMGCTDPRP 331
S+ST S SNG A + S+ VFA LFASSTAS S LQ Q PAFSDLIR+MG +
Sbjct: 167 GSNSTCSTSNGGATSNSNSSSSVFAGLFASSTASGS--LQSQTPAFSDLIRAMGPPEHPA 340
Query: 332 TDFSAPPSSEAISLCLSTSHGSSIFGTGGQECRQYVPTHQPPAMSATALLQKAAQMGAAA 391
SAP SSEAISLCLST+ S IF TGG RQY + PAMSATALLQKAAQMGAAA
Sbjct: 341 DLISAPSSSEAISLCLSTTSASPIFATGG---RQYASSSPHPAMSATALLQKAAQMGAAA 511
Query: 392 TNASLLRGLGIVSSSA--STSSGQQDSLHWG-LGPMEPEGSSLVSAGLGLGLPC-DADSG 447
TNASLLRGLGIVSSS STSSGQQ+ L WG +EPE S+ +AGLGLGLPC D SG
Sbjct: 512 TNASLLRGLGIVSSSPLDSTSSGQQNVLQWGHQQQLEPESGSVAAAGLGLGLPCEDGGSG 691
Query: 448 LKELMLGTPSMFGPKQTTLDFLGLGMAAGGSAGGGLSALITSIGGGSGLDVTAAAASFGN 507
LKELM+GT SMFGPK TTLDFLGLGMAAGG+AGGGLSALITSIGG GLDVTAA FGN
Sbjct: 692 LKELMMGTSSMFGPKHTTLDFLGLGMAAGGTAGGGLSALITSIGG--GLDVTAA---FGN 856
Query: 508 GEFSGKDIGRSS 519
G+FSGKDIG SS
Sbjct: 857 GDFSGKDIGSSS 892
>BI245412 homologue to GP|29028906|gb At2g02080 {Arabidopsis thaliana},
partial (31%)
Length = 664
Score = 257 bits (657), Expect = 8e-69
Identities = 115/174 (66%), Positives = 141/174 (80%), Gaps = 4/174 (2%)
Frame = -1
Query: 26 SPIPKPTKKKRNLPGMPDPEAEVIALSPTTLL----ATNRFVCEICNKGFQRDQNLQLHR 81
+P+P P +K+ PE + +T+ + +C KGFQR+QNLQLHR
Sbjct: 661 TPVPPPQRKEN------PPEHHIQCGGDSTISQDPNGNKQVYM*VCTKGFQREQNLQLHR 500
Query: 82 RGHNLPWKLRQRSSKEIRKRVYVCPEPTCVHHDPSRALGDLTGIKKHFCRKHGEKKWKCE 141
RGHNLPWKL+Q+++KE +++VY+CPEPTCVHHDPSRALGDLTGIKKH+ RKHGEKKWKC+
Sbjct: 499 RGHNLPWKLKQKTTKEPKRKVYLCPEPTCVHHDPSRALGDLTGIKKHYSRKHGEKKWKCD 320
Query: 142 KCSKKYAVQSDWKAHSKVCGSREYKCDCGTVFSRRDSFITHRAFCDALAEENAK 195
KCSKKYAVQSDWKAHSK CG+REY+CDCGT+FSRRDSFITHRAFCDALA+E+A+
Sbjct: 319 KCSKKYAVQSDWKAHSKTCGTREYRCDCGTLFSRRDSFITHRAFCDALAQESAR 158
Score = 47.8 bits (112), Expect = 1e-05
Identities = 29/81 (35%), Positives = 42/81 (51%)
Frame = -2
Query: 25 QSPIPKPTKKKRNLPGMPDPEAEVIALSPTTLLATNRFVCEICNKGFQRDQNLQLHRRGH 84
Q P P +K+ +L + + AEVIALSP TL+ATNRF+C KG + +
Sbjct: 663 QHQCPPPKEKRTHLNTISN--AEVIALSPKTLMATNRFICRCAPKGSKESKTYSYTEEDT 490
Query: 85 NLPWKLRQRSSKEIRKRVYVC 105
L +R K R+R ++C
Sbjct: 489 TCLGSLSRRLQKSQRER-FIC 430
>BG362868 homologue to PIR|T46147|T461 zinc finger protein - Arabidopsis
thaliana, partial (21%)
Length = 482
Score = 250 bits (638), Expect = 1e-66
Identities = 120/154 (77%), Positives = 132/154 (84%), Gaps = 4/154 (2%)
Frame = +1
Query: 98 IRKRVYVCPEPTCVHHDPSRALGDLTGIKKHFCRKHGEKKWKCEKCSKKYAVQSDWKAHS 157
+RKRVYVCPEPTCVHHDPSRALGDLTGIKKHFCRKHGEKKWKC+KCSKKYAV SDWKAHS
Sbjct: 16 VRKRVYVCPEPTCVHHDPSRALGDLTGIKKHFCRKHGEKKWKCDKCSKKYAVHSDWKAHS 195
Query: 158 KVCGSREYKCDCGTVFSRRDSFITHRAFCDALAEENAKSQNQAVGKANSESDSKVLTGDS 217
K+CG+REYKCDCGT+FSRRDSFITHRAFCDALAEE+A+SQ Q V KA+SESDSK +TGDS
Sbjct: 196 KICGTREYKCDCGTLFSRRDSFITHRAFCDALAEESARSQPQTVAKASSESDSKAVTGDS 375
Query: 218 LP--AVITTTAAAAATTPQSNSGV--SSALETQK 247
P AV + QSNS V SS+L+TQK
Sbjct: 376 SPPVAVEAPPPLVPPVSSQSNSVVVPSSSLQTQK 477
>BQ080665
Length = 425
Score = 238 bits (608), Expect = 4e-63
Identities = 104/121 (85%), Positives = 116/121 (94%)
Frame = -2
Query: 43 DPEAEVIALSPTTLLATNRFVCEICNKGFQRDQNLQLHRRGHNLPWKLRQRSSKEIRKRV 102
DP AEVIALSP TL+ATNRFVCEICNKGFQRDQNLQLHRRGHNLPWKL+ R++ ++RKRV
Sbjct: 364 DPSAEVIALSPNTLVATNRFVCEICNKGFQRDQNLQLHRRGHNLPWKLKLRTTTDVRKRV 185
Query: 103 YVCPEPTCVHHDPSRALGDLTGIKKHFCRKHGEKKWKCEKCSKKYAVQSDWKAHSKVCGS 162
YVCPEP+CVHH+P+RALGDLTGIKKHF RKHGEKKWKCEKCSKKYAVQSDWKAHSK+CG+
Sbjct: 184 YVCPEPSCVHHNPARALGDLTGIKKHFSRKHGEKKWKCEKCSKKYAVQSDWKAHSKICGT 5
Query: 163 R 163
+
Sbjct: 4 K 2
>TC222859 homologue to GB|AAO64832.1|29028906|BT005897 At2g02080 {Arabidopsis
thaliana;} , partial (22%)
Length = 454
Score = 228 bits (580), Expect = 6e-60
Identities = 98/121 (80%), Positives = 114/121 (93%)
Frame = +2
Query: 37 NLPGMPDPEAEVIALSPTTLLATNRFVCEICNKGFQRDQNLQLHRRGHNLPWKLRQRSSK 96
N+ DP+AEVIALSP TL+ATNRF+CE+CNKGFQR+QNLQLHRRGHNLPWKL+Q+++K
Sbjct: 92 NICAYADPDAEVIALSPKTLMATNRFLCEVCNKGFQREQNLQLHRRGHNLPWKLKQKTNK 271
Query: 97 EIRKRVYVCPEPTCVHHDPSRALGDLTGIKKHFCRKHGEKKWKCEKCSKKYAVQSDWKAH 156
E +++VY+CPEPTCVHHDPSRALGDLTGIKKH+ RKHGEKKWKC+KCSKKYAVQSDWKAH
Sbjct: 272 EPKRKVYLCPEPTCVHHDPSRALGDLTGIKKHYSRKHGEKKWKCDKCSKKYAVQSDWKAH 451
Query: 157 S 157
S
Sbjct: 452 S 454
>CA938031
Length = 416
Score = 201 bits (512), Expect = 5e-52
Identities = 96/130 (73%), Positives = 112/130 (85%), Gaps = 8/130 (6%)
Frame = +1
Query: 3 DLDNVSTASGEASISSSGNN-----NIQSPIPKPT---KKKRNLPGMPDPEAEVIALSPT 54
++ N+++ASGEAS +SSGN + +P P T KKKRNLPG PDP+AEVIALSP
Sbjct: 25 NMSNLTSASGEASAASSGNRTEIGTSYMAPPPSQTQQSKKKRNLPGNPDPDAEVIALSPK 204
Query: 55 TLLATNRFVCEICNKGFQRDQNLQLHRRGHNLPWKLRQRSSKEIRKRVYVCPEPTCVHHD 114
+LLATNRF+CEICNKGFQRDQNLQLHRRGHNLPWKL+QR+SKE+RK+VYVCPEP+CVHHD
Sbjct: 205 SLLATNRFICEICNKGFQRDQNLQLHRRGHNLPWKLKQRTSKEVRKKVYVCPEPSCVHHD 384
Query: 115 PSRALGDLTG 124
PSRALGDLTG
Sbjct: 385 PSRALGDLTG 414
>BF069705 similar to GP|16226322|gb AT3g45260/F18N11_20 {Arabidopsis
thaliana}, partial (22%)
Length = 406
Score = 200 bits (508), Expect = 1e-51
Identities = 86/120 (71%), Positives = 104/120 (86%)
Frame = +1
Query: 94 SSKEIRKRVYVCPEPTCVHHDPSRALGDLTGIKKHFCRKHGEKKWKCEKCSKKYAVQSDW 153
S+KE+RK+VY+CPE TCVHHD +RALGDLTGIKKH+ RKHGEKKWKCEKCSKKYAVQSDW
Sbjct: 4 SNKEVRKKVYICPEKTCVHHDAARALGDLTGIKKHYSRKHGEKKWKCEKCSKKYAVQSDW 183
Query: 154 KAHSKVCGSREYKCDCGTVFSRRDSFITHRAFCDALAEENAKSQNQAVGKANSESDSKVL 213
KAH+K CG+REYKCDCG +FSR+DSFITHRAFCDALA+E+++ + A N +S+ +
Sbjct: 184 KAHTKTCGTREYKCDCGNLFSRKDSFITHRAFCDALADESSRLTSVASTSLNFKSEDATM 363
>TC233614
Length = 460
Score = 190 bits (483), Expect = 1e-48
Identities = 90/143 (62%), Positives = 109/143 (75%), Gaps = 3/143 (2%)
Frame = +1
Query: 65 EICNKGFQRDQNLQLHRRGHNLPWKLRQRSSKEIRKRVYVCPEPTCVHHDPSRALGDLTG 124
EICN+GFQRDQNLQ+HRR H +PWKL +R + ++KRV+VCPEP+C+HHDP ALGDL G
Sbjct: 1 EICNQGFQRDQNLQMHRRRHKVPWKLLKRETPVVKKRVFVCPEPSCLHHDPCHALGDLVG 180
Query: 125 IKKHFCRKH-GEKKWKCEKCSKKYAVQSDWKAHSKVCGSREYKCDCGTVFSRRDSFITHR 183
IKKHF RKH K+W CE+CSK YAVQSD+KAH K CG+R + CDCG VFSR +SFI H+
Sbjct: 181 IKKHFRRKHNNHKQWVCERCSKGYAVQSDYKAHLKTCGTRGHSCDCGRVFSRVESFIEHQ 360
Query: 184 AFCDA--LAEENAKSQNQAVGKA 204
C+ L E +SQ QAV A
Sbjct: 361 DACNVGRLGPE-TQSQAQAVQAA 426
>CA936809 homologue to PIR|T46147|T461 zinc finger protein - Arabidopsis
thaliana, partial (14%)
Length = 423
Score = 184 bits (466), Expect = 1e-46
Identities = 99/142 (69%), Positives = 106/142 (73%), Gaps = 15/142 (10%)
Frame = +3
Query: 131 RKHGEKKWKCEKCSKKYAVQSDWKAHSKVCGSREYKCDCGTVFSRRDSFITHRAFCDALA 190
RKHGEKKWKC+KCSKKYAVQSDWKAHSKVCG+REYKCDCGTVFSRRDSFITHRAFCDALA
Sbjct: 3 RKHGEKKWKCDKCSKKYAVQSDWKAHSKVCGTREYKCDCGTVFSRRDSFITHRAFCDALA 182
Query: 191 EENAKSQNQAVGKANSESDSKVLTGDSLP------AVITTTAAAAAT--------TPQSN 236
EEN S V K SE+DSKVL GDS P V TTAAA T T Q+N
Sbjct: 183 EENFLS--HTVVKDISENDSKVLIGDSPPPQPVAATVAATTAAATTTTTTTTITSTTQAN 356
Query: 237 SGVSSALETQKLDLPE-NPPQI 257
S +SS L+T LDLPE NPPQ+
Sbjct: 357 SVMSSCLQTHNLDLPENNPPQV 422
>BU084794
Length = 420
Score = 182 bits (462), Expect = 3e-46
Identities = 79/101 (78%), Positives = 90/101 (88%)
Frame = +2
Query: 109 TCVHHDPSRALGDLTGIKKHFCRKHGEKKWKCEKCSKKYAVQSDWKAHSKVCGSREYKCD 168
+CVHHDPSRALGDLTGIKKH+ RKHGEKKWKC+KCSKKYAVQSDWKAHSK+CG+REYKCD
Sbjct: 2 SCVHHDPSRALGDLTGIKKHYSRKHGEKKWKCDKCSKKYAVQSDWKAHSKICGTREYKCD 181
Query: 169 CGTVFSRRDSFITHRAFCDALAEENAKSQNQAVGKANSESD 209
CGT+FSR+DSFITHRAFCDALAEE+A+ +N +D
Sbjct: 182 CGTLFSRKDSFITHRAFCDALAEESARVTTVPAALSNLRND 304
>TC211735
Length = 689
Score = 178 bits (451), Expect = 6e-45
Identities = 88/128 (68%), Positives = 98/128 (75%), Gaps = 14/128 (10%)
Frame = +1
Query: 4 LDNVSTASGEASISSSGNNNIQSPI-------------PKPTKKKRNLPGMPDPEAEVIA 50
+ N+++ASGEAS SS I + P P KKKRNLPG PDPEAEV+A
Sbjct: 304 MSNLTSASGEASASSGNRTEIGTDYSQQYFAPPLSQAQPPPLKKKRNLPGNPDPEAEVVA 483
Query: 51 LSPTTLLATNRFVCEICNKGFQRDQNLQLHRRGHNLPWKLRQRSSKE-IRKRVYVCPEPT 109
LSP TLLATNRF+CEICNKGFQRDQNLQLHRRGHNLPWKL+QRSS E IRK+VYVCPE +
Sbjct: 484 LSPKTLLATNRFICEICNKGFQRDQNLQLHRRGHNLPWKLKQRSSNEIIRKKVYVCPEAS 663
Query: 110 CVHHDPSR 117
CVHHDPSR
Sbjct: 664 CVHHDPSR 687
>TC214744 homologue to UP|Q9LVQ7 (Q9LVQ7) Zinc finger protein, partial (19%)
Length = 1045
Score = 167 bits (424), Expect = 8e-42
Identities = 111/274 (40%), Positives = 134/274 (48%)
Frame = +1
Query: 122 LTGIKKHFCRKHGEKKWKCEKCSKKYAVQSDWKAHSKVCGSREYKCDCGTVFSRRDSFIT 181
LTGIKKHFCRKHGEKKWKC+KCSKKYAVQSDWKAHSK+CG+REYKCDCGT+FSRRDSFIT
Sbjct: 1 LTGIKKHFCRKHGEKKWKCDKCSKKYAVQSDWKAHSKICGTREYKCDCGTIFSRRDSFIT 180
Query: 182 HRAFCDALAEENAKSQNQAVGKANSESDSKVLTGDSLPAVITTTAAAAATTPQSNSGVSS 241
HRAFCDALAEEN K+ + K L +P ++++
Sbjct: 181 HRAFCDALAEENNKANEGQLPKIGPN-----LQCQQIPNLVSS----------------- 294
Query: 242 ALETQKLDLPENPPQIIEEPQVVVTTTASALNASCSSSTSSKSNGCAATSTGVFASLFAS 301
LP N I+ P + TT+ +A S + F + A+
Sbjct: 295 --------LPINTNSIVPNPAQMGGTTSEFNHADHKHPLSLPHELMPMPAQKPFNNNMAA 450
Query: 302 STASASASLQPQAPAFSDLIRSMGCTDPRPTDFSAPPSSEAISLCLSTSHGSSIFGTGGQ 361
T RS+ T SS ++ L S++F G
Sbjct: 451 GTV---------------FTRSLSST-----------SSPSLQL------SSNMFDENGL 534
Query: 362 ECRQYVPTHQPPAMSATALLQKAAQMGAAATNAS 395
P MSATALLQKAAQMGA T S
Sbjct: 535 HLAA-----GSPHMSATALLQKAAQMGATLTEKS 621
>BE660007 similar to GP|29028906|gb At2g02080 {Arabidopsis thaliana}, partial
(25%)
Length = 667
Score = 147 bits (372), Expect = 9e-36
Identities = 71/122 (58%), Positives = 90/122 (73%), Gaps = 1/122 (0%)
Frame = +2
Query: 26 SPIPKPTKKKRNLPGMPDPEAEVIALSPTTLLATNRFVCEICNKGFQRDQNLQLHRRGHN 85
SP P KK+RN PG P P+AEVI LSP TL+ATNRF+CE+CNKGFQR+QNLQLHRRGHN
Sbjct: 236 SPTTAPQKKRRNQPGTPYPDAEVIKLSPKTLMATNRFICEVCNKGFQREQNLQLHRRGHN 415
Query: 86 LPWKLRQRS-SKEIRKRVYVCPEPTCVHHDPSRALGDLTGIKKHFCRKHGEKKWKCEKCS 144
LPWKL+Q+S +KE +++VY+CP P + L + + + + EKKWKCEKCS
Sbjct: 416 LPWKLKQKSTTKEPKRKVYLCPSPP-ASIMILQGLWETSLGSRSLLSQAXEKKWKCEKCS 592
Query: 145 KK 146
K+
Sbjct: 593 KE 598
Score = 43.5 bits (101), Expect = 2e-04
Identities = 25/80 (31%), Positives = 39/80 (48%), Gaps = 3/80 (3%)
Frame = +3
Query: 92 QRSSKEIRKRVYVCPEPTCVHHDPS---RALGDLTGIKKHFCRKHGEKKWKCEKCSKKYA 148
+R ++ +K ++C H PS + G ++ + RK + + +KYA
Sbjct: 435 RRVQQKSQKERFICVR---AHLRPS*SFKGFGRPHWDQEAYYRKPARRNGSVKSAQRKYA 605
Query: 149 VQSDWKAHSKVCGSREYKCD 168
QSDWKAHSK RE+ CD
Sbjct: 606 GQSDWKAHSKTLWPREFXCD 665
>TC230714 weakly similar to GB|AAO44076.1|28466935|BT004810 At5g22890
{Arabidopsis thaliana;} , partial (58%)
Length = 1049
Score = 109 bits (272), Expect = 3e-24
Identities = 70/203 (34%), Positives = 101/203 (49%), Gaps = 18/203 (8%)
Frame = +1
Query: 25 QSPIPKPT---KKKRNLPGMPDP----EAEVIALSPTTLLATNRFVCEICNKGFQRDQNL 77
+ P P+P+ K K+ L +P ++E++ L +LA + CEIC KGF+RD NL
Sbjct: 187 KKPKPEPSVADKAKQTLDSKLEPLEGDDSEIVELDAVEILAEHMHFCEICGKGFRRDSNL 366
Query: 78 QLHRRGHNLPWKLRQRSSK------EIRKRVYVCPEPTCVH---HDPSRALGDLTGIKKH 128
++H R H +K + +K + R + CP C H R L + +K H
Sbjct: 367 RMHMRAHGEQFKTVEALAKPSETTAQRRATRFSCPFEGCNRNKLHRRFRPLKSVICVKNH 546
Query: 129 FCRKHGEKKWKCEKCSKK-YAVQSDWKAHSKVCGSR-EYKCDCGTVFSRRDSFITHRAFC 186
F R H K + CE+C KK ++V SD ++H+K CG +KC CGT FSR+D H A
Sbjct: 547 FKRSHCPKMYTCERCRKKHFSVLSDLRSHAKHCGGEARWKCTCGTTFSRKDKLFGHIALF 726
Query: 187 DALAEENAKSQNQAVGKANSESD 209
D A A + GK E D
Sbjct: 727 DGHAPALA-CDEEGKGKQVVEDD 792
>TC206813 similar to UP|CW14_YEAST (O13547) Covalently-linked cell wall
protein 14 precursor (Inner cell wall protein), partial
(6%)
Length = 1214
Score = 106 bits (264), Expect = 3e-23
Identities = 57/176 (32%), Positives = 87/176 (49%), Gaps = 12/176 (6%)
Frame = +1
Query: 21 NNNIQSPIPKPTKKKRNLPGMPDPEAEVIALSPTTLLATNRFVCEICNKGFQRDQNLQLH 80
N N++ PK + +P E++ L +LA + C IC KGF+RD NL++H
Sbjct: 643 NYNMEEHEPKDEEDADEGENLPPGSYEILQLEKEEILAPHTHFCTICGKGFKRDANLRMH 822
Query: 81 RRGHNLPWKL--------RQRSSKEIRKRVYVCPEPTCVH---HDPSRALGDLTGIKKHF 129
RGH +K ++ S+ + Y CP C H + L + +K H+
Sbjct: 823 MRGHGDKYKTPAALAKPHKETGSEPKLIKRYSCPYAGCKRNKDHKKFQPLKTILCVKNHY 1002
Query: 130 CRKHGEKKWKCEKC-SKKYAVQSDWKAHSKVCGSREYKCDCGTVFSRRDSFITHRA 184
R H +K + C +C +KK++V +D K H K CG ++ C CGT FSR+D H A
Sbjct: 1003KRTHCDKSYTCSRCNTKKFSVMADLKTHEKHCGKDKWLCSCGTTFSRKDKLFGHIA 1170
>TC224111 similar to UP|Q944L3 (Q944L3) AT3g45260/F18N11_20 (ID1-like zinc
finger protein 1), partial (17%)
Length = 688
Score = 105 bits (263), Expect = 4e-23
Identities = 47/79 (59%), Positives = 63/79 (79%), Gaps = 4/79 (5%)
Frame = +2
Query: 23 NIQSPIPKPT----KKKRNLPGMPDPEAEVIALSPTTLLATNRFVCEICNKGFQRDQNLQ 78
++Q P P P K++R+LP PDP+AEV+ALSP L+ATNRF+ E+C++G QRDQNLQ
Sbjct: 437 HVQPPSPYPNSNAAKRRRSLPRTPDPDAEVVALSPKCLMATNRFLWEVCSEGVQRDQNLQ 616
Query: 79 LHRRGHNLPWKLRQRSSKE 97
LHRRGHNLPW+LR+R+ +
Sbjct: 617 LHRRGHNLPWRLRKRTDND 673
>CA801722
Length = 451
Score = 102 bits (253), Expect = 5e-22
Identities = 70/168 (41%), Positives = 94/168 (55%), Gaps = 5/168 (2%)
Frame = +3
Query: 162 SREYKCDCGTVFSRRDSFITHRAFCDALAEENAKSQNQAVGKANSESDSKVLTGDSLPAV 221
SREY+CDCGT+FSRRDSFITHRAFCD LA+E+A++Q QA G+ S+SKV+ S P
Sbjct: 3 SREYRCDCGTLFSRRDSFITHRAFCDVLAQESARAQAQAQGQGG--SNSKVVEASSPPTP 176
Query: 222 ITTTAAAAATTPQSNSGVSSALETQKLDLPENPPQIIEEPQVVVTTTASALNASCSSSTS 281
T +A S V L Q ++PENP + + A+A + SSST
Sbjct: 177 PLTPSA---------SVVPPTLSIQSSEIPENPSTKLSP----TSPNATACFVTSSSSTP 317
Query: 282 SKSNGCAATSTGVFASLFASSTASASASLQPQAP-----AFSDLIRSM 324
S S C T VFAS+ A S + PQ+P +F++LI ++
Sbjct: 318 SNSTTC----TTVFASVLAPSPTT------PQSPSNSTSSFTNLISNL 431
>TC231421 homologue to GB|AAM10292.1|20147151|AY091693 AT3g13810/MCP4_2
{Arabidopsis thaliana;} , partial (12%)
Length = 1164
Score = 99.0 bits (245), Expect = 5e-21
Identities = 45/60 (75%), Positives = 52/60 (86%), Gaps = 1/60 (1%)
Frame = +3
Query: 148 AVQSDWKAHSKVCGSREYKCDCGTVFSRRDSFITHRAFCDALAEENAK-SQNQAVGKANS 206
AVQSDWKAHSK CG+REY+CDCGT+FSR+DSFITHRAFCDALAEE+A+ S NQ A +
Sbjct: 3 AVQSDWKAHSKTCGTREYRCDCGTLFSRKDSFITHRAFCDALAEESARLSANQLAAAAGA 182
>BM527349
Length = 408
Score = 89.7 bits (221), Expect = 3e-18
Identities = 44/84 (52%), Positives = 61/84 (72%)
Frame = +1
Query: 1 MVDLDNVSTASGEASISSSGNNNIQSPIPKPTKKKRNLPGMPDPEAEVIALSPTTLLATN 60
M+D +N ++ SG S S + ++ + K+KR G PDP+AEV++LSPTTLL ++
Sbjct: 160 MLDNNNSASNSGAPSSSDAAFALSENGVAN-NKRKRRPAGTPDPDAEVVSLSPTTLLESD 336
Query: 61 RFVCEICNKGFQRDQNLQLHRRGH 84
R+VCEICN+GFQRDQNLQ+HRR H
Sbjct: 337 RYVCEICNQGFQRDQNLQMHRRRH 408
>AW780545
Length = 409
Score = 89.4 bits (220), Expect = 4e-18
Identities = 36/46 (78%), Positives = 44/46 (95%)
Frame = +2
Query: 150 QSDWKAHSKVCGSREYKCDCGTVFSRRDSFITHRAFCDALAEENAK 195
++DWKAHSK CG+REY+CDCGT+FSRRDSFITHRAFCDALA ++A+
Sbjct: 2 RADWKAHSKTCGTREYRCDCGTLFSRRDSFITHRAFCDALAHDSAR 139
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.312 0.127 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,321,323
Number of Sequences: 63676
Number of extensions: 414216
Number of successful extensions: 3364
Number of sequences better than 10.0: 233
Number of HSP's better than 10.0 without gapping: 3035
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3270
length of query: 519
length of database: 12,639,632
effective HSP length: 102
effective length of query: 417
effective length of database: 6,144,680
effective search space: 2562331560
effective search space used: 2562331560
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC148484.5


