

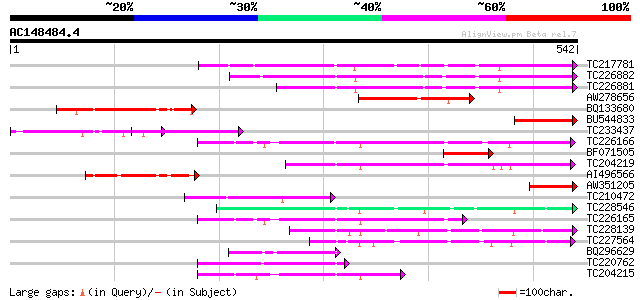
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148484.4 - phase: 0
(542 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC217781 similar to UP|O81316 (O81316) F6N15.20 protein (Scarecr... 197 8e-51
TC226882 similar to UP|O81316 (O81316) F6N15.20 protein (Scarecr... 180 1e-45
TC226881 similar to UP|O81316 (O81316) F6N15.20 protein (Scarecr... 157 9e-39
AW278656 similar to GP|9800311|gb|A pr95.1 {rat cytomegalovirus ... 149 4e-36
BQ133680 127 2e-29
BU544833 similar to GP|26452022|dbj ap2 SCARECROW-like protein {... 119 4e-27
TC233437 similar to UP|Q9FYW2 (Q9FYW2) BAC19.14, partial (10%) 107 1e-23
TC226166 similar to UP|O23724 (O23724) GAI protein, partial (70%) 97 1e-20
BF071505 similar to GP|26452022|dbj ap2 SCARECROW-like protein {... 93 3e-19
TC204219 similar to UP|Q7Y1B6 (Q7Y1B6) GAI-like protein, partial... 93 3e-19
AI496566 87 1e-17
AW351205 similar to GP|9858782|gb|A BAC19.14 {Lycopersicon escul... 87 2e-17
TC210472 similar to UP|Q9FHZ1 (Q9FHZ1) SCARECROW gene regulator-... 69 7e-12
TC228546 similar to UP|Q8W584 (Q8W584) AT4g17230/dl4650c, partia... 68 1e-11
TC226165 GAI1 [Glycine max] 66 5e-11
TC228139 similar to UP|Q8GVE1 (Q8GVE1) Chitin-inducible gibberel... 63 4e-10
TC227564 similar to UP|Q9LDL7 (Q9LDL7) SCARECROW gene regulator-... 61 1e-09
BQ296629 similar to GP|17064924|gb SCARECROW gene regulator {Ara... 59 7e-09
TC220762 similar to UP|Q6S5L6 (Q6S5L6) GAI protein, partial (27%) 58 1e-08
TC204215 similar to UP|Q84TQ7 (Q84TQ7) GIA/RGA-like gibberellin ... 56 5e-08
>TC217781 similar to UP|O81316 (O81316) F6N15.20 protein (Scarecrow-like 6)
(SCL6) (AT4g00150/F6N15_20), partial (42%)
Length = 1762
Score = 197 bits (502), Expect = 8e-51
Identities = 133/368 (36%), Positives = 202/368 (54%), Gaps = 6/368 (1%)
Frame = +1
Query: 181 EELIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFHFKDALQSLLSGSNRTNP 240
E+L +AA+ D +L+LA IL RLN +L SP GKP RAAF+FK+ALQ LL N N
Sbjct: 421 EQLFKAAELIDAGNLELAHGILARLNHQL-SPIGKPFQRAAFYFKEALQLLLH-PNANNS 594
Query: 241 PRLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGSLYMHVVDFEIGLGIQY 300
S ++ I +K+FS ISP+ F+ FT NQALLEA+ G +H++DF+IGLG Q+
Sbjct: 595 SFTFSPTGLLLKIGAYKSFSEISPVLQFANFTCNQALLEAVEGFDRIHIIDFDIGLGGQW 774
Query: 301 ASLMKEIAEKAVNGS-PLLRITAVVPEEY--AVESRLIRENLNQFAHDLGIRVQVDFVPL 357
+S M+E+A + NGS P L+ITA V + +E +E+L Q+A +L + +++ + L
Sbjct: 775 SSFMQELALR--NGSAPELKITAFVSPSHHDEIELSFSQESLKQYAGELHMSFELEILSL 948
Query: 358 RTFETVSFKAVRFVDGEKTAILLTPAIFCRLGSEGTAAFLSDVRRITPGVVVFVDGEGWT 417
+ + S+ D E + + F S L V+++ P +VV +D
Sbjct: 949 ESLNSASWPQP-LRDCEAVVVNMPIGSFSNYPSY-LPLVLRFVKQLMPKIVVTLDRSC-- 1116
Query: 418 EAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWARRIEMLLLRP---KIIAAVEA 474
A F + ++ +L+ YS +LESLDA + + IE L+P K++
Sbjct: 1117-DRTDAPFPQHLIFALQSYSGLLESLDA---VNVHPDVLQMIEKYYLQPSMEKLVLGRHG 1284
Query: 475 AGRRTTPWREAFYGAGMRPVQLSQFADFQAECLLAKVQIRGFHVAKRQAELVLFWHERAM 534
R PW+ +G P+ S F + QAECL+ + +GFHV KRQ+ LVL W + +
Sbjct: 1285LQERALPWKNLLLSSGFSPLTFSNFTESQAECLVQRTPSKGFHVEKRQSSLVLCWQRKDL 1464
Query: 535 VATSAWRC 542
++ S WRC
Sbjct: 1465ISVSTWRC 1488
>TC226882 similar to UP|O81316 (O81316) F6N15.20 protein (Scarecrow-like 6)
(SCL6) (AT4g00150/F6N15_20), partial (32%)
Length = 1423
Score = 180 bits (457), Expect = 1e-45
Identities = 116/338 (34%), Positives = 184/338 (54%), Gaps = 6/338 (1%)
Frame = +3
Query: 211 SPTGKPLHRAAFHFKDALQSLLSGSNRTNPPRLSSMVEIVQTIRTFKAFSGISPIPMFSI 270
SP G+P RAAF+ K+AL SLL + + S + + I +K+FS ISP+ F+
Sbjct: 9 SPIGRPFQRAAFYMKEALMSLLHSNAHSF--MAFSPISFIFKIGAYKSFSEISPVLQFAN 182
Query: 271 FTTNQALLEALHGSLYMHVVDFEIGLGIQYASLMKEIAEKAVNGSPLLRITAVVPEEYA- 329
FT NQAL+EA+ S +HV+DF+IG G+Q++S M+EIA ++ +G+P L++TA+V
Sbjct: 183 FTCNQALIEAVERSDRIHVIDFDIGFGVQWSSFMQEIALRS-SGAPSLKVTAIVSPSTCD 359
Query: 330 -VESRLIRENLNQFAHDLGIRVQVDFVPLRTFETVSFKAV-RFVDGEKTAILLTPAIFCR 387
VE RENL Q+A D+ + + + + + + + S + +F D E + + + F
Sbjct: 360 EVELNFTRENLIQYAKDINVSFEFNVLSIESLNSPSCPLLGKFFDNEAIVVNMPVSSFTN 539
Query: 388 LGSEGTAAFLSDVRRITPGVVVFVDGEGWTEAAAAASFRRGVVNSLEFYSMMLESLDASV 447
S + L V+++ P VVV +D VV+ L+ YS +LESLDA
Sbjct: 540 YPSLFPSV-LHFVKQLRPKVVVTLDR---ICDQMDVPLPTNVVHVLQCYSALLESLDA-- 701
Query: 448 AAGGGGEWARRIEMLLLRP---KIIAAVEAAGRRTTPWREAFYGAGMRPVQLSQFADFQA 504
+ ++IE ++P KII + PWR F +G P S F + QA
Sbjct: 702 -VNVNLDVLQKIERHFIQPAIKKIILGHHHFQEKLPPWRNLFMQSGFSPFTFSNFTEAQA 878
Query: 505 ECLLAKVQIRGFHVAKRQAELVLFWHERAMVATSAWRC 542
ECL+ + +RGFHV ++ + LVL W ++ +++ S WRC
Sbjct: 879 ECLVQRAPVRGFHVERKPSSLVLCWQKKELISVSTWRC 992
>TC226881 similar to UP|O81316 (O81316) F6N15.20 protein (Scarecrow-like 6)
(SCL6) (AT4g00150/F6N15_20), partial (26%)
Length = 871
Score = 157 bits (398), Expect = 9e-39
Identities = 99/293 (33%), Positives = 161/293 (54%), Gaps = 6/293 (2%)
Frame = +2
Query: 256 FKAFSGISPIPMFSIFTTNQALLEALHGSLYMHVVDFEIGLGIQYASLMKEIAEKAVNGS 315
+K+FS ISP+ F+ FT NQAL+EA+ +HV+DF+IG G+Q++S M+E+A ++ +G+
Sbjct: 2 YKSFSEISPVLQFANFTCNQALIEAVERFDRIHVIDFDIGFGVQWSSFMQELALRS-SGA 178
Query: 316 PLLRITAVVPEEYA--VESRLIRENLNQFAHDLGIRVQVDFVPLRTFETVSFKAV-RFVD 372
P L++TA+V VE RENL Q+A D+ + +++ + + + S + +F D
Sbjct: 179 PSLKVTAIVSPSTCDEVELNFTRENLIQYAKDINVSFELNVFSIESLNSASCPLLGQFFD 358
Query: 373 GEKTAILLTPAIFCRLGSEGTAAFLSDVRRITPGVVVFVDGEGWTEAAAAASFRRGVVNS 432
E A+ + + F S + L V+++ P VVV +D VV+
Sbjct: 359 NEAIAVNMPVSSFTNYPSLFPSV-LHFVKQLRPKVVVTLDR---ICDRIDVPLPTNVVHV 526
Query: 433 LEFYSMMLESLDASVAAGGGGEWARRIEMLLLRP---KIIAAVEAAGRRTTPWREAFYGA 489
L+ YS +LESLDA + ++IE ++P KII + + PWR F +
Sbjct: 527 LQCYSALLESLDA---VNVNLDALQKIERHFIQPAIKKIILGHHHSQEKLPPWRNLFIQS 697
Query: 490 GMRPVQLSQFADFQAECLLAKVQIRGFHVAKRQAELVLFWHERAMVATSAWRC 542
G P S F + QAECL+ + +RGFHV ++ + LVL W + +++ S WRC
Sbjct: 698 GFSPFTFSNFTEAQAECLVQRAPVRGFHVERKPSSLVLCWQRKELISVSTWRC 856
>AW278656 similar to GP|9800311|gb|A pr95.1 {rat cytomegalovirus Maastricht}
[Rat cytomegalovirus Maastricht], partial (2%)
Length = 341
Score = 149 bits (375), Expect = 4e-36
Identities = 78/113 (69%), Positives = 95/113 (84%), Gaps = 2/113 (1%)
Frame = +2
Query: 334 LIRENLNQFAHDLGIRVQVDFVPLRTFETVSFKAVRFVDGEKTAILLTPAIFCRLGSEGT 393
L+ +NL QFA +L IRVQV+FV LRTFE +SFK+++FVDGE T +LL+PAIF LG+
Sbjct: 8 LVHDNLAQFALELRIRVQVEFVALRTFENLSFKSLKFVDGENTTVLLSPAIFGHLGN--A 181
Query: 394 AAFLSDVRRITPGVVVFVDGEGWTE--AAAAASFRRGVVNSLEFYSMMLESLD 444
AAFL+DVRRI+P +VVFVDGEGW E A+AASFRRGVV+SLE+YSMMLESLD
Sbjct: 182 AAFLADVRRISPSMVVFVDGEGWAETATASAASFRRGVVSSLEYYSMMLESLD 340
>BQ133680
Length = 421
Score = 127 bits (318), Expect = 2e-29
Identities = 71/145 (48%), Positives = 89/145 (60%), Gaps = 11/145 (7%)
Frame = +2
Query: 45 YEPRSVLELCRSPSPEKK-------PSQQEQEHNPEVEQEDHASLPNLDWWDSIMKDLGL 97
YEP SVL+LCRSPSPEKK P +N ++ +DH LPN DWW+SIMKDL L
Sbjct: 2 YEPTSVLDLCRSPSPEKKLTVPKPEPQHNNNNNNLNLDLDDHV-LPNSDWWESIMKDLAL 178
Query: 98 QDDSSTPIIPLLKNTNNTSEIYPNPSQDQFDQTQDFTSLSDIYSNNQNLAYNYPNTNTNL 157
+DS TP++ N + + P+ FD QDFTSLS+IY NQNL YNYP TNT L
Sbjct: 179 PEDSPTPLLKTNINPSCIPDFPPSSQDPPFDHPQDFTSLSEIY--NQNLPYNYP-TNT-L 346
Query: 158 DHLVNDFNNNQPNNN----TNSNWD 178
+H D ++N +NN N+NWD
Sbjct: 347 EHSFYDLSHNHHHNNNNNVNNNNWD 421
>BU544833 similar to GP|26452022|dbj ap2 SCARECROW-like protein {Arabidopsis
thaliana}, partial (12%)
Length = 394
Score = 119 bits (297), Expect = 4e-27
Identities = 56/60 (93%), Positives = 58/60 (96%)
Frame = -1
Query: 483 REAFYGAGMRPVQLSQFADFQAECLLAKVQIRGFHVAKRQAELVLFWHERAMVATSAWRC 542
REAFYGAGMRPVQLSQFADFQAECLLAK QIRGFHVAKRQ ELVLFWH+RA+VATSAWRC
Sbjct: 394 REAFYGAGMRPVQLSQFADFQAECLLAKSQIRGFHVAKRQNELVLFWHDRAIVATSAWRC 215
>TC233437 similar to UP|Q9FYW2 (Q9FYW2) BAC19.14, partial (10%)
Length = 727
Score = 107 bits (268), Expect = 1e-23
Identities = 74/161 (45%), Positives = 93/161 (56%), Gaps = 11/161 (6%)
Frame = +2
Query: 1 MRVPPPAVSSSSSSPQPKLTTTQNIVVNVPTPNPTPTPTTTT-LCYEPRSVLELCRSPSP 59
MRVP P SSPQ T N+V + PN + T T LCYEP SVL+LCRSPSP
Sbjct: 86 MRVPVP------SSPQANPKPTNNVVRTIALPNLNNSTTPPTGLCYEPTSVLDLCRSPSP 247
Query: 60 -EKKPSQQEQ----EHNPEVEQEDHASLPNLDWWDSIMKDLGLQDDSSTPIIP--LLKNT 112
+KP+ ++ EDHA L NLD WDSIMKDLGL DDS+TP++ L +
Sbjct: 248 GTEKPTTDHSVLVTNSQDYLDLEDHA-LHNLD-WDSIMKDLGLHDDSATPVLKTFLHPDD 421
Query: 113 NNTSEIYPNPSQDQ---FDQTQDFTSLSDIYSNNQNLAYNY 150
++ + NPS D FD +FT+LSDIYS NQN + +
Sbjct: 422 DDDDDNNNNPSCDDFTLFDHALEFTTLSDIYS-NQNFGFRF 541
Score = 88.2 bits (217), Expect = 8e-18
Identities = 49/107 (45%), Positives = 63/107 (58%)
Frame = +3
Query: 117 EIYPNPSQDQFDQTQDFTSLSDIYSNNQNLAYNYPNTNTNLDHLVNDFNNNQPNNNTNSN 176
+++ P+ T + +S+ L T+T LV DFN+ + N +
Sbjct: 399 KLFSTPTTTTTTTTTTTHHATTSHSSTTRLNSPLSPTSTPTKTLVFDFNHLPHDFNHLNG 578
Query: 177 WDFIEELIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFH 223
+DFIEELIRA DCFD L +AQ ILERLNQRLRSP GKPLHRAAF+
Sbjct: 579 FDFIEELIRAVDCFDTKQLHVAQLILERLNQRLRSPVGKPLHRAAFY 719
>TC226166 similar to UP|O23724 (O23724) GAI protein, partial (70%)
Length = 1613
Score = 97.4 bits (241), Expect = 1e-20
Identities = 96/375 (25%), Positives = 159/375 (41%), Gaps = 13/375 (3%)
Frame = +1
Query: 180 IEELIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFHFKDALQSLLSGSNRTN 239
+ L+ A+ +NN+L +A+A+++++ S G + + A +F +AL + R
Sbjct: 115 VHSLMACAEAVENNNLAVAEALVKQIGFLAVSQVGA-MRKVAIYFAEALARRIY---RVF 282
Query: 240 PPR--LSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGSLYMHVVDFEIGLG 297
P + LS ++I F P F+ FT NQ +LEA G +HV+DF I G
Sbjct: 283 PLQHSLSDSLQI--------HFYETCPYLKFAHFTANQVILEAFQGKNRVHVIDFGINQG 438
Query: 298 IQYASLMKEIAEKAVNGSPLLRITAVVPEEYAVESRL--IRENLNQFAHDLGIRVQVDFV 355
+Q+ +LM+ +A + G P+ R+T + P L + L Q A ++ ++ +
Sbjct: 439 MQWPALMQALAVR-TGGPPVFRLTGIGPPAADNSDHLQEVGWKLAQLAEEINVQFEYRGF 615
Query: 356 PLRTFETVSFKAVRFVDGEKTAILLTPAIFCRLGSEGTA-AFLSDVRRITPGVVVFVDGE 414
+ + + + E A+ L G LS VR+I P ++ V+ E
Sbjct: 616 VANSLADLDASMLDLREDESVAVNSVFEFHKLLARPGAVEKVLSVVRQIRPEILTVVEQE 795
Query: 415 GWTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWARRIEMLLLRPKIIAAVEA 474
SF SL +YS + +SL+ S + + + L +I V
Sbjct: 796 A---NHNGLSFVDRFTESLHYYSTLFDSLEGSPVNPND----KAMSEVYLGKQICNVVAC 954
Query: 475 AG-------RRTTPWREAFYGAGMRPVQLSQFADFQAECLLAKV-QIRGFHVAKRQAELV 526
G WR F G PV L A QA LL+ G+ V + L+
Sbjct: 955 EGMDRVERHETLNQWRNRFGSTGFSPVHLGSNAYKQASMLLSLFGGGDGYRVEENNGCLM 1134
Query: 527 LFWHERAMVATSAWR 541
L WH R ++ATS W+
Sbjct: 1135LGWHTRPLIATSVWQ 1179
>BF071505 similar to GP|26452022|dbj ap2 SCARECROW-like protein {Arabidopsis
thaliana}, partial (6%)
Length = 148
Score = 93.2 bits (230), Expect = 3e-19
Identities = 45/48 (93%), Positives = 47/48 (97%)
Frame = +3
Query: 415 GWTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWARRIEML 462
GWTEAAAAASFRRGVV+SLEFYSMMLESLDASVA+GGGGEW RRIEML
Sbjct: 3 GWTEAAAAASFRRGVVSSLEFYSMMLESLDASVASGGGGEWVRRIEML 146
>TC204219 similar to UP|Q7Y1B6 (Q7Y1B6) GAI-like protein, partial (50%)
Length = 1208
Score = 92.8 bits (229), Expect = 3e-19
Identities = 84/291 (28%), Positives = 133/291 (44%), Gaps = 13/291 (4%)
Frame = +1
Query: 264 PIPMFSIFTTNQALLEALHGSLYMHVVDFEIGLGIQYASLMKEIAEKAVNGSPLLRITAV 323
P F+ FT NQA+LEA + +HV+DF + G+Q+ +LM+ +A + G P R+T +
Sbjct: 31 PYLKFAHFTANQAILEAFATAGKVHVIDFGLKQGMQWPALMQALALRP-GGPPTFRLTGI 207
Query: 324 VPEEYAVESRL--IRENLNQFAHDLGIRVQVDFVPLRTFETVSFKAVRFVDGEKTAILLT 381
P + L + L Q A ++G++ + + + K + GE A+
Sbjct: 208 GPPQPDNTDALQQVGWKLAQLAQNIGVQFEFRGFVCNSLADLDPKMLEIRPGEAVAVNSV 387
Query: 382 PAIFCRLGSEGTA-AFLSDVRRITPGVVVFVDGEGWTEAAAAASFRRGVVNSLEFYSMML 440
+ L G+ L V++I P +V V+ E F +L +YS +
Sbjct: 388 FELHRMLARPGSVDKVLDTVKKIKPKIVTIVEQEA---NHNGPGFLDRFTEALHYYSSLF 558
Query: 441 ESLDASVAAGGGGEWARRIEM--LLLRPKI--IAAVEAAGR-----RTTPWREAFYGAGM 491
+SL+ S ++ G G + + M L L +I + A E A R + WR AG
Sbjct: 559 DSLEGSSSSTGLGSPNQDLLMSELYLGRQICNVVANEGADRVERHETLSQWRGRLDSAGF 738
Query: 492 RPVQLSQFADFQAECLLAK-VQIRGFHVAKRQAELVLFWHERAMVATSAWR 541
PV L A QA LLA G+ V + L+L WH R ++ATSAW+
Sbjct: 739 DPVHLGSNAFKQASMLLALFAGGDGYRVEENNGCLMLGWHTRPLIATSAWK 891
>AI496566
Length = 312
Score = 87.4 bits (215), Expect = 1e-17
Identities = 55/112 (49%), Positives = 72/112 (64%), Gaps = 3/112 (2%)
Frame = +1
Query: 73 EVEQEDHASLPNLDWWDSIMKDLGLQDDSSTPIIPLLKNTNNTSEIYPNPSQD-QFDQ-T 130
+++ +DH LPN DWW+SIMKDL L +DS T PLLK + +P SQD FD T
Sbjct: 10 QLDLDDHV-LPNSDWWESIMKDLTLPEDSPT---PLLKTNPSCIPDFPPSSQDPSFDHPT 177
Query: 131 QDFTSLSDIYSNNQNLAYNYPNTNTNLDHLVNDFNNN-QPNNNTNSNWDFIE 181
DFTSLS+IY NQN+ +NYP + L+ +D N++ P+NN NWDFIE
Sbjct: 178 PDFTSLSEIY--NQNIPFNYP--SNTLEPSFHDLNHSLHPHNN---NWDFIE 312
>AW351205 similar to GP|9858782|gb|A BAC19.14 {Lycopersicon esculentum},
partial (8%)
Length = 447
Score = 87.0 bits (214), Expect = 2e-17
Identities = 40/45 (88%), Positives = 42/45 (92%)
Frame = -1
Query: 498 QFADFQAECLLAKVQIRGFHVAKRQAELVLFWHERAMVATSAWRC 542
QFAD+QAECLLAKVQIRGFHV KR AELVL WHERAMV+TSAWRC
Sbjct: 447 QFADYQAECLLAKVQIRGFHVDKRHAELVLCWHERAMVSTSAWRC 313
>TC210472 similar to UP|Q9FHZ1 (Q9FHZ1) SCARECROW gene regulator-like
protein, partial (44%)
Length = 751
Score = 68.6 bits (166), Expect = 7e-12
Identities = 42/148 (28%), Positives = 78/148 (52%), Gaps = 4/148 (2%)
Frame = +1
Query: 168 QPNNNTNSNWDFIEELIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFHFKDA 227
+P+ ++ + L++ A+C ++L A +L + + L SP G R +F A
Sbjct: 160 EPHGGDSTGLKLLGLLLQCAECVAMDNLDFANDLLPEIAE-LSSPYGTSPERVGAYFAQA 336
Query: 228 LQS-LLSGSNRTNPPRLSSMVEIVQTIRTFKAF---SGISPIPMFSIFTTNQALLEALHG 283
LQ+ ++S + P + V + Q+ + F AF + +SP+ FS FT NQA+ +AL G
Sbjct: 337 LQARVVSSCIGSYSPLTAKSVTLTQSQKIFNAFQSYNSVSPLVKFSHFTANQAIFQALDG 516
Query: 284 SLYMHVVDFEIGLGIQYASLMKEIAEKA 311
+H++D +I G+Q+ L +A ++
Sbjct: 517 EDRVHIIDLDIMQGLQWPGLFHILASRS 600
>TC228546 similar to UP|Q8W584 (Q8W584) AT4g17230/dl4650c, partial (60%)
Length = 1493
Score = 67.8 bits (164), Expect = 1e-11
Identities = 78/364 (21%), Positives = 140/364 (38%), Gaps = 19/364 (5%)
Frame = +3
Query: 198 AQAILERLNQRLRSPTGKPLHRAAFHFKDALQSLLSGSNRTNPPRLSSMVEIVQTIRTFK 257
A + + ++ S G P+ R + + L++ L S L + T+
Sbjct: 6 AVGFMNNVLAKMVSVGGDPIQRLGAYMLEGLRARLESSGSIIYKALKCEQPTSNDLMTYM 185
Query: 258 -AFSGISPIPMFSIFTTNQALLEALHGSLYMHVVDFEIGLGIQYASLMKEIAEKAVNGSP 316
I P F+ + N + EA+ + ++DF+I G Q+ L++ +A + G P
Sbjct: 186 HILYQICPYWKFAYTSANAVIGEAMLNESRIRIIDFQIAQGTQWLLLIQALASRP-GGPP 362
Query: 317 LLRITAVVPEEYAVESR-----LIRENLNQFAHDLGIRVQVDFVPLRTFETVSFKAVRFV 371
+ +T V + + +R ++ + L+ +A G+ + + E V +
Sbjct: 363 FVHVTGV-DDSQSFHARGGGLHIVGKRLSDYAKSCGVPFEFHSAAMCGSELELENLV--I 533
Query: 372 DGEKTAILLTPAIFCRLGSEGTAA------FLSDVRRITPGVVVFVDGEGWTEAAAAASF 425
+ ++ P + + E + L V+ ++P VV V+ E T + F
Sbjct: 534 QPGEALVVNFPFVLHHMPDESVSTENHRDRLLRLVKSLSPKVVTLVEQESNTNTSP---F 704
Query: 426 RRGVVNSLEFYSMMLESLDASVAAGGGGEWARRIEMLLLRPKIIAAVEAAGRRTTP---- 481
+ V +L +Y+ M ES+D VA + E + I+ V G
Sbjct: 705 FQRFVETLSYYTAMFESID--VALPRDDKQRINAEQHCVARDIVNMVACEGDERLERHEL 878
Query: 482 ---WREAFYGAGMRPVQLSQFADFQAECLLAKVQIRGFHVAKRQAELVLFWHERAMVATS 538
WR F AG P LS +L + + + R L L W RAM +S
Sbjct: 879 LGKWRSRFSMAGFAPCPLSSSVTAAVRNMLNEFN-ENYRLQHRDGALYLGWKSRAMCTSS 1055
Query: 539 AWRC 542
AWRC
Sbjct: 1056AWRC 1067
>TC226165 GAI1 [Glycine max]
Length = 1313
Score = 65.9 bits (159), Expect = 5e-11
Identities = 67/263 (25%), Positives = 116/263 (43%), Gaps = 5/263 (1%)
Frame = +3
Query: 180 IEELIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFHFKDALQSLLSGSNRTN 239
+ L+ A+ +NN+L +A+A+++++ S G + + A +F +AL + R
Sbjct: 570 VHSLMACAEAVENNNLAVAEALVKQIGFLALSQVGA-MRKVATYFAEALARRIY---RVF 737
Query: 240 PPR--LSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGSLYMHVVDFEIGLG 297
P + LS ++I F P F+ FT NQA+LEA G +HV+DF I G
Sbjct: 738 PQQHSLSDSLQI--------HFYETCPYLKFAHFTANQAILEAFQGKNRVHVIDFGINQG 893
Query: 298 IQYASLMKEIAEKAVNGSPLLRITAVVPEEYAVESRL--IRENLNQFAHDLGIRVQVDFV 355
+Q+ +LM+ +A + +G P+ R+T + P L + L Q A + ++ +
Sbjct: 894 MQWPALMQALALRN-DGPPVFRLTGIGPPAADNSDHLQEVGWKLAQLAERIHVQFEYRGF 1070
Query: 356 PLRTFETVSFKAVRFVDGEKTAILLTPAIFCRLGSEGTA-AFLSDVRRITPGVVVFVDGE 414
+ + + + E A+ L G LS VR+I P ++ V+ E
Sbjct: 1071VANSLADLDASMLDLREDESVAVNSVFEFHKLLARPGAVEKVLSVVRQIRPEILTVVEQE 1250
Query: 415 GWTEAAAAASFRRGVVNSLEFYS 437
SF SL +YS
Sbjct: 1251A---NHNGLSFVDRFTESLHYYS 1310
>TC228139 similar to UP|Q8GVE1 (Q8GVE1) Chitin-inducible
gibberellin-responsive protein, partial (47%)
Length = 1497
Score = 62.8 bits (151), Expect = 4e-10
Identities = 63/291 (21%), Positives = 120/291 (40%), Gaps = 16/291 (5%)
Frame = +1
Query: 268 FSIFTTNQALLEALHGSLYMHVVDFEIGLGIQYASLMKEIAEKAVNGSPLLRITAV--VP 325
F N A+ +A ++H++DF+I G Q+ +L++ +A + G+P +RIT +
Sbjct: 13 FGYMAANGAIAQACRNEDHIHIIDFQIAQGTQWMTLLQALAARP-GGAPHVRITGIDDPV 189
Query: 326 EEYAVESRL--IRENLNQFAHDLGIRVQVDFVPLRTFETVSFKAVRFVDGEKTAILLTPA 383
+YA L + + L + GI V+ VP+ V+ + + GE A+
Sbjct: 190 SKYARGDGLEVVGKRLALMSEKFGIPVEFHGVPVFA-PNVTREMLDIRPGEALAVNFPLQ 366
Query: 384 IFCRLG-----SEGTAAFLSDVRRITPGVVVFVDGEGWTEAAAAASFRRGVVNSLEFYSM 438
+ S L VR ++P V V+ E T F + +L++Y
Sbjct: 367 LHHTADESVHVSNPRDGLLRLVRSLSPKVTTLVEQESNTN---TTPFFNRFIETLDYYLA 537
Query: 439 MLESLDASVAAGGGGEWARRIEMLLLRPKIIAAVEAAGRRTT-------PWREAFYGAGM 491
+ ES+D ++ + +E L I+ + G+ W+ AG
Sbjct: 538 IFESIDVTLPR--DSKERINVEQHCLARDIVNIIACEGKERVERHELFGKWKSRLKMAGF 711
Query: 492 RPVQLSQFADFQAECLLAKVQIRGFHVAKRQAELVLFWHERAMVATSAWRC 542
+ LS + + LL + + ++ ++L W +R +++ SAW C
Sbjct: 712 QQCPLSSYVNSVIRSLL-MCYSEHYTLVEKDGAMLLGWKDRNLISASAWHC 861
>TC227564 similar to UP|Q9LDL7 (Q9LDL7) SCARECROW gene regulator-like
(Phytochrome A signal transduction 1 protein), partial
(53%)
Length = 1107
Score = 61.2 bits (147), Expect = 1e-09
Identities = 72/277 (25%), Positives = 117/277 (41%), Gaps = 22/277 (7%)
Frame = +3
Query: 287 MHVVDFEIGLGIQYASLMKEIAEKAVNGSPLLRITAVVPEEYAVESR-----LIRENLNQ 341
+H+VDF+IG G Q+ SL++ +A + V G P +RI+ V + Y+ +R ++ + L+
Sbjct: 6 VHIVDFQIGQGTQWVSLIQALARRPV-GPPKIRISG-VDDSYSAYARRGGLDIVGKRLSA 179
Query: 342 FAHDL-------GIRVQVDFVPLRTFETVSFKAVRFVDGEKTAILLTPAIFCRLGSEG-T 393
A +RV V V L E ++AV AI L + S
Sbjct: 180 LAQSCHVPFEFNAVRVPVTEVQLEDLELRPYEAV----AVNFAISLHHVPDESVNSHNHR 347
Query: 394 AAFLSDVRRITPGVVVFVDGEGWTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGG 453
L ++++P VV V+ E T A F + V ++ +Y + ES+D +
Sbjct: 348 DRLLRLAKQLSPKVVTLVEQEFSTN---NAPFLQRFVETMNYYLAVFESIDTVLPR---- 506
Query: 454 EWARRI--EMLLLRPKIIAAVEAAGRR-------TTPWREAFYGAGMRPVQLSQFADFQA 504
E RI E L +++ + G WR F AG P LS +
Sbjct: 507 EHKERINVEQHCLAREVVNLIACEGEERVERHELLNKWRMRFTKAGFTPYPLSSVINSSI 686
Query: 505 ECLLAKVQIRGFHVAKRQAELVLFWHERAMVATSAWR 541
+ LL + + +R L L W + +VA+ AWR
Sbjct: 687 KDLLQSYH-GHYTLEERDGALFLGWMNQVLVASCAWR 794
>BQ296629 similar to GP|17064924|gb SCARECROW gene regulator {Arabidopsis
thaliana}, partial (19%)
Length = 437
Score = 58.5 bits (140), Expect = 7e-09
Identities = 38/107 (35%), Positives = 56/107 (51%)
Frame = +1
Query: 210 RSPTGKPLHRAAFHFKDALQSLLSGSNRTNPPRLSSMVEIVQTIRTFKAFSGISPIPMFS 269
R P KP R F+F AL + G P SS E+ + +KA + P F+
Sbjct: 1 RLPARKPPERVGFYFWQALSRKMWGDKEKMEP--SSWEELTLS---YKALNDACPYSKFA 165
Query: 270 IFTTNQALLEALHGSLYMHVVDFEIGLGIQYASLMKEIAEKAVNGSP 316
T NQA+LEA + +H++DF I GIQ+A+L++ A +A +G P
Sbjct: 166 HLTANQAILEATENASNIHILDFGIVQGIQWAALLQAFATRA-SGKP 303
>TC220762 similar to UP|Q6S5L6 (Q6S5L6) GAI protein, partial (27%)
Length = 725
Score = 57.8 bits (138), Expect = 1e-08
Identities = 43/147 (29%), Positives = 72/147 (48%), Gaps = 1/147 (0%)
Frame = +1
Query: 180 IEELIRAADCFDNNHLQLAQAILERLNQRLRSP-TGKPLHRAAFHFKDALQSLLSGSNRT 238
+ L+ AD L A +++E + L T + + A +F DAL+ + G
Sbjct: 49 VHTLMTCADSVQRGDLAFAGSLIENMQGLLAHVNTNIGIGKVAGYFIDALRRRILGQGVF 228
Query: 239 NPPRLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGSLYMHVVDFEIGLGI 298
LSS + + + P F+ FT NQA+LEA +G +HV+DF + G+
Sbjct: 229 QT--LSSSSYPYEDNVLYHHYYEACPYLKFAHFTANQAILEAFNGHDCVHVIDFNLMQGL 402
Query: 299 QYASLMKEIAEKAVNGSPLLRITAVVP 325
Q+ +L++ +A + G PLLR+T + P
Sbjct: 403 QWPALIQALALRP-GGPPLLRLTGIGP 480
>TC204215 similar to UP|Q84TQ7 (Q84TQ7) GIA/RGA-like gibberellin response
modulator, partial (45%)
Length = 1589
Score = 55.8 bits (133), Expect = 5e-08
Identities = 49/203 (24%), Positives = 90/203 (44%), Gaps = 4/203 (1%)
Frame = +1
Query: 180 IEELIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFHFKDALQSLLSG--SNR 237
+ L+ A+ +L+LA A+++ + S G + + A +F AL + G
Sbjct: 736 VHTLLACAEAVQQENLKLADALVKHVGILAASQAGA-MRKVASYFAQALARRIYGIFPEE 912
Query: 238 TNPPRLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGSLYMHVVDFEIGLG 297
T S ++ + F P F+ FT NQA+LEA + +HV+DF + G
Sbjct: 913 TLDSSFSDVLHM--------HFYESCPYLKFAHFTANQAILEAFATAGRVHVIDFGLRQG 1068
Query: 298 IQYASLMKEIAEKAVNGSPLLRITAVVPEEYAVESRL--IRENLNQFAHDLGIRVQVDFV 355
+Q+ +LM+ +A + G P R+T + P + L + L Q A ++G++ +
Sbjct: 1069MQWPALMQALALRP-GGPPTFRLTGIGPPQPDNTDALQQVGWKLAQLAQNIGVQFEFRGF 1245
Query: 356 PLRTFETVSFKAVRFVDGEKTAI 378
+ + K + GE A+
Sbjct: 1246VCNSLADLDPKMLEIRPGEAVAV 1314
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.133 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,636,691
Number of Sequences: 63676
Number of extensions: 405966
Number of successful extensions: 6013
Number of sequences better than 10.0: 288
Number of HSP's better than 10.0 without gapping: 4304
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5307
length of query: 542
length of database: 12,639,632
effective HSP length: 102
effective length of query: 440
effective length of database: 6,144,680
effective search space: 2703659200
effective search space used: 2703659200
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC148484.4


