

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148484.3 + phase: 0 /pseudo
(341 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
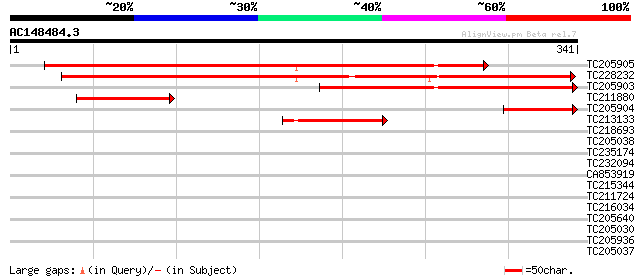
Score E
Sequences producing significant alignments: (bits) Value
TC205905 similar to UP|Q41113 (Q41113) BZIP transcriptional repr... 280 7e-76
TC228232 homologue to UP|Q43449 (Q43449) G-box binding factor, c... 263 6e-71
TC205903 similar to UP|Q41113 (Q41113) BZIP transcriptional repr... 186 2e-47
TC211880 homologue to UP|Q43449 (Q43449) G-box binding factor, p... 66 3e-11
TC205904 similar to UP|Q41113 (Q41113) BZIP transcriptional repr... 64 1e-10
TC213133 homologue to UP|Q43449 (Q43449) G-box binding factor, p... 63 2e-10
TC218693 similar to UP|Q94KA8 (Q94KA8) BZIP transcription factor... 34 0.11
TC205038 similar to UP|Q94II3 (Q94II3) ERD3 protein, partial (48%) 32 0.54
TC235174 31 0.71
TC232094 homologue to UP|Q9S834 (Q9S834) ATP-dependent Clp prote... 30 1.6
CA853919 homologue to PIR|T11622|T11 extensin class 1 precursor ... 29 2.7
TC215344 homologue to UP|Q8LP18 (Q8LP18) Cullin-like protein1, p... 28 4.6
TC211724 similar to UP|Q8LLE5 (Q8LLE5) Coiled-coil protein (Frag... 28 6.0
TC216034 28 6.0
TC205640 similar to UP|Q9FHW0 (Q9FHW0) Gb|AAF07835.1 (At5g37070)... 28 6.0
TC205030 UP|ACCD_SOYBN (P49158) Acetyl-coenzyme A carboxylase ca... 28 6.0
TC205936 similar to GB|AAL58101.1|18056655|AF395058 CSN complex ... 28 7.8
TC205037 similar to UP|Q94II3 (Q94II3) ERD3 protein, partial (83%) 28 7.8
>TC205905 similar to UP|Q41113 (Q41113) BZIP transcriptional repressor ROM1,
partial (83%)
Length = 1226
Score = 280 bits (716), Expect = 7e-76
Identities = 171/269 (63%), Positives = 191/269 (70%), Gaps = 2/269 (0%)
Frame = +2
Query: 22 RKYQHQQYNHHIQIGQPPCRPTIILEPLRLPIMPQLWLHQPRILICGEASIL*WHHTGLQ 81
R+ + Q + HI GQ PCRP ++LE L LP +PQL L Q I I GEASIL* H GLQ
Sbjct: 425 RQLRRHQQHLHILTGQAPCRPIMLLEALHLPFLPQLLLPQLPIPIYGEASIL*CRHMGLQ 604
Query: 82 FRILQCSLLGISMLILAW**LQVLCTKLQSLKGRDLMERIRIHLKNRRALLQIQAPKQER 141
I LLG+SMLILAW *+QVL +K+Q LKGR+L E I H KN + LLQIQ PKQE+
Sbjct: 605 SHIQLYILLGVSMLILAWQ*IQVLSSKIQRLKGRELKENIGTHPKN*KDLLQIQLPKQEK 784
Query: 142 VERQDQVQAMMAFHIVVTVVQRVHLMLAMK--TNRNQLETRREALTSCLLMEPTRRTILL 199
VERQ QVQAMMA+ VV VVQRVH MLAM+ TNRNQL+ RREALT CLLMEP RTILL
Sbjct: 785 VERQAQVQAMMAYRKVVKVVQRVHQMLAMRILTNRNQLQIRREALT*CLLMEPMHRTILL 964
Query: 200 DPFLNHLFLEILLSRYLQLILILEWTYGMHLLLVLKPPK*DTINLVLLELVHLVNSGCNK 259
FLN L LE LLS+ QLIL LEWT GMHLL+ LK K*D INLV EL ++N K
Sbjct: 965 VLFLNLLCLESLLSQCQQLILTLEWTCGMHLLVALKLQK*DIINLVPRELPLVING--YK 1138
Query: 260 MIVS*KDRRENSLIESQPGGQDYASRPSV 288
M VS*KDR NS ES GGQDYASR SV
Sbjct: 1139MNVS*KDR*GNSRTESHLGGQDYASRLSV 1225
>TC228232 homologue to UP|Q43449 (Q43449) G-box binding factor, complete
Length = 1723
Score = 263 bits (673), Expect = 6e-71
Identities = 176/319 (55%), Positives = 208/319 (65%), Gaps = 10/319 (3%)
Frame = +2
Query: 32 HIQIGQPPCRPTIILEPLRLPIMPQLWLHQPRILICGEASIL*WHHTGLQFRILQCSLLG 91
HI IGQ CR T++LEPL L +PQ+ L Q ILICGEAS L*+HH L F I LLG
Sbjct: 116 HILIGQARCRLTMLLEPLHLHFLPQILLLQLPILICGEASTL*FHHIVLLFHIQLYILLG 295
Query: 92 ISMLILAW**LQVLCTKLQSLKGRDLMERIRIHLKNRRALLQIQAPKQERVERQDQVQAM 151
+SMLILAW * +QSL R LM++I K +R LQ PK E +ERQ QV M
Sbjct: 296 MSMLILAWQ*P*APHRMVQSL*ERVLMKKIGFLPKVQRLCLQTMVPKLETMERQAQVPEM 475
Query: 152 MAFHIVVTVVQRVHLMLAMK--TNRNQLETRREALTSCLLMEPTRRTILLDPFLNHLFLE 209
MA H V+ VVQR ML M+ TNRN+L+TR+ LT CLLM TIL FLN LE
Sbjct: 476 MAPHKVLKVVQRDLRMLVMRILTNRNRLQTRKGVLTKCLLMVLMHGTIL*ASFLN---LE 646
Query: 210 ILLSRYLQLILILEWTYGMHLLLVLKPPK*DTINLVLLELVH--------LVNSGCNKMI 261
I L + LQL+LILE T GMHLLLV K K*D I+L L L+ VN+G KM
Sbjct: 647 IPLCQCLQLVLILE*TCGMHLLLVTKLQK*DRISLQELLLLQP*WDVKSRWVNTGY-KMN 823
Query: 262 VS*KDRRENSLIESQPGGQDYASRPSVKNYKRGWRRWEVRIELSEKSFRNFLKSVRSLHL 321
V+*++R+ NSLI S G QDYASR SVK+YKRGW WEVRI+LSEKSFR +LK+ ++LHL
Sbjct: 824 VN*RNRKGNSLIGSLLGDQDYASRLSVKSYKRGWSLWEVRIKLSEKSFREYLKNAKNLHL 1003
Query: 322 KTILLRKTWNGCVGLK*LL 340
K I RK+ NGCV K LL
Sbjct: 1004KMIPSRKS*NGCVDQKQLL 1060
>TC205903 similar to UP|Q41113 (Q41113) BZIP transcriptional repressor ROM1,
partial (46%)
Length = 827
Score = 186 bits (471), Expect = 2e-47
Identities = 106/155 (68%), Positives = 115/155 (73%)
Frame = +2
Query: 187 CLLMEPTRRTILLDPFLNHLFLEILLSRYLQLILILEWTYGMHLLLVLKPPK*DTINLVL 246
CLLME RTILL FLN L LE L QLIL LEWT GMHLL+ LK K*D INLV
Sbjct: 5 CLLMEXMXRTILLVLFLNXLCLESLCXXCQQLILTLEWTCGMHLLVALKLQK*DIINLVP 184
Query: 247 LELVHLVNSGCNKMIVS*KDRRENSLIESQPGGQDYASRPSVKNYKRGWRRWEVRIELSE 306
EL ++N KM VS*KDRR NS ESQPGGQDYASR SVK+YKRGW RWEVRI+LSE
Sbjct: 185 RELPLVING--YKMNVS*KDRRGNSQTESQPGGQDYASRLSVKSYKRGWNRWEVRIKLSE 358
Query: 307 KSFRNFLKSVRSLHLKTILLRKTWNGCVGLK*LLT 341
KSFR+FLK+ RSLH K IL RK+W+GCV K LLT
Sbjct: 359 KSFRDFLKNARSLHPKIILSRKSWSGCVVQKQLLT 463
>TC211880 homologue to UP|Q43449 (Q43449) G-box binding factor, partial (24%)
Length = 726
Score = 65.9 bits (159), Expect = 3e-11
Identities = 36/59 (61%), Positives = 42/59 (71%)
Frame = +1
Query: 41 RPTIILEPLRLPIMPQLWLHQPRILICGEASIL*WHHTGLQFRILQCSLLGISMLILAW 99
R T++LEPL L +PQ+ L Q ILICGEAS L*+HH L F I LLG+SMLILAW
Sbjct: 505 RLTMLLEPLHLHFLPQILLLQLPILICGEASTL*FHHIVLLFHIQLYILLGMSMLILAW 681
>TC205904 similar to UP|Q41113 (Q41113) BZIP transcriptional repressor ROM1,
partial (26%)
Length = 730
Score = 63.9 bits (154), Expect = 1e-10
Identities = 31/44 (70%), Positives = 36/44 (81%)
Frame = +1
Query: 298 WEVRIELSEKSFRNFLKSVRSLHLKTILLRKTWNGCVGLK*LLT 341
WEVRI+LSE SFR+FLK+ RSLH K IL RK+W+GCV K LLT
Sbjct: 262 WEVRIKLSEMSFRDFLKNARSLHQKIILSRKSWSGCVVQKQLLT 393
>TC213133 homologue to UP|Q43449 (Q43449) G-box binding factor, partial (21%)
Length = 718
Score = 62.8 bits (151), Expect = 2e-10
Identities = 36/63 (57%), Positives = 43/63 (68%)
Frame = +3
Query: 165 HLMLAMKTNRNQLETRREALTSCLLMEPTRRTILLDPFLNHLFLEILLSRYLQLILILEW 224
HL++ + RNQL+TR+ LT CLLMEP IL PFL LEI LS+ LQL+LILEW
Sbjct: 156 HLLVLLL--RNQLQTRKGVLTKCLLMEPMHGRIL*APFLILQCLEIPLSQCLQLVLILEW 329
Query: 225 TYG 227
T G
Sbjct: 330 TCG 338
Score = 28.5 bits (62), Expect = 4.6
Identities = 24/74 (32%), Positives = 29/74 (38%), Gaps = 16/74 (21%)
Frame = +2
Query: 1 MGTKEDSTTKPSKTSSSTQ-----TNRKYQHQQY-----------NHHIQIGQPPCRPTI 44
MGT E+ST K SK SSS+ TN+K Q I P P +
Sbjct: 116 MGTDEESTAKVSKPSSSSSTQESATNKKGSFDQMLVDGANARKNSVSTIPHSSVPGNPAV 295
Query: 45 ILEPLRLPIMPQLW 58
+ P L I LW
Sbjct: 296 SMSPTSLNIGMDLW 337
>TC218693 similar to UP|Q94KA8 (Q94KA8) BZIP transcription factor 2, partial
(61%)
Length = 1226
Score = 33.9 bits (76), Expect = 0.11
Identities = 47/181 (25%), Positives = 82/181 (44%), Gaps = 15/181 (8%)
Frame = +1
Query: 125 LKNRRALLQIQAPKQERVERQDQVQAMMAFHIVVTVVQRVHLMLAMKTNRNQLETRREAL 184
+K++ LL PK +V+ + V+ +M I++ +K N ++ + L
Sbjct: 10 VKHQENLLMGFIPKVVKVQVTELVKEVMRTPIMILN*NPGRGRTLLKMNHLKMAVQYMLL 189
Query: 185 TSCLLMEPTRRTILLDPFLNHLFL-EILLSRYL--QLILILEWTYGMHLLLVLKPPK*DT 241
+++ R +L+ L++L+L ++L+ ++L QLI I EW G H L+PP
Sbjct: 190 KMGYIID--LRQLLIKLCLSYLYLPQVLVEQFLVPQLI*I*EWIIGAH---QLRPPFLHC 354
Query: 242 INLVLL------------ELVHLVNSGCNKMIVS*KDRRENSLIESQPGGQDYASRPSVK 289
+ LL E+V GC + S KD+ +SLI + Y SR +V
Sbjct: 355 MERFLLLQLLEG*LLLDHEMVFSHKFGC-RTKESLKDKGGSSLIGNLHADPGYGSRLNVM 531
Query: 290 N 290
N
Sbjct: 532 N 534
>TC205038 similar to UP|Q94II3 (Q94II3) ERD3 protein, partial (48%)
Length = 889
Score = 31.6 bits (70), Expect = 0.54
Identities = 16/47 (34%), Positives = 23/47 (48%)
Frame = +2
Query: 276 QPGGQDYASRPSVKNYKRGWRRWEVRIELSEKSFRNFLKSVRSLHLK 322
+PGG S P + NY+R WR W IE + + + + SL K
Sbjct: 587 RPGGFWVLSGPPI-NYERRWRGWNTTIEAQKSDYEKLKELLTSLCFK 724
>TC235174
Length = 711
Score = 31.2 bits (69), Expect = 0.71
Identities = 24/87 (27%), Positives = 38/87 (43%)
Frame = -1
Query: 155 HIVVTVVQRVHLMLAMKTNRNQLETRREALTSCLLMEPTRRTILLDPFLNHLFLEILLSR 214
H+V + VHL L T R + +TRR A T + RRT++ P + + L
Sbjct: 258 HLVCNSLVAVHLPLGFATRRTKPQTRRNATTLHTWVCTHRRTLVSFP----VVVSSPLLL 91
Query: 215 YLQLILILEWTYGMHLLLVLKPPK*DT 241
+L L + W + + + *DT
Sbjct: 90 HLAPTLFVAWIFSVMEFTLYSEAS*DT 10
>TC232094 homologue to UP|Q9S834 (Q9S834) ATP-dependent Clp protease subunit
ClpP (NClpP1) (ATP-dependent Clp protease proteolytic
subunit ClpP5) , partial (49%)
Length = 842
Score = 30.0 bits (66), Expect = 1.6
Identities = 16/40 (40%), Positives = 27/40 (67%)
Frame = +3
Query: 83 RILQCSLLGISMLILAW**LQVLCTKLQSLKGRDLMERIR 122
R+L CS+ G+ ++IL * ++VL +Q LK RD+ +I+
Sbjct: 588 RVLFCSVQGLKVMILLP*AVRVLFCSVQGLKERDIACQIQ 707
>CA853919 homologue to PIR|T11622|T11 extensin class 1 precursor - cowpea,
partial (30%)
Length = 454
Score = 29.3 bits (64), Expect = 2.7
Identities = 19/55 (34%), Positives = 23/55 (41%)
Frame = +1
Query: 26 HQQYNHHIQIGQPPCRPTIILEPLRLPIMPQLWLHQPRILICGEASIL*WHHTGL 80
H NHH + P TII P LPI L H P +L+ + HH L
Sbjct: 127 HTIINHHPHLHHPLLLLTIINLPHHLPITINLLHHHPPLLLPPTTTTPHHHHLPL 291
>TC215344 homologue to UP|Q8LP18 (Q8LP18) Cullin-like protein1, partial (92%)
Length = 2465
Score = 28.5 bits (62), Expect = 4.6
Identities = 14/45 (31%), Positives = 26/45 (57%)
Frame = +2
Query: 208 LEILLSRYLQLILILEWTYGMHLLLVLKPPK*DTINLVLLELVHL 252
+E+ Y +L LEW + + ++LKPP T+ L++LE + +
Sbjct: 326 IELY*RMY*TYLLKLEWGKWITMRMILKPPCSKTLLLIILERLRI 460
>TC211724 similar to UP|Q8LLE5 (Q8LLE5) Coiled-coil protein (Fragment),
partial (12%)
Length = 1086
Score = 28.1 bits (61), Expect = 6.0
Identities = 14/41 (34%), Positives = 24/41 (58%)
Frame = +1
Query: 290 NYKRGWRRWEVRIELSEKSFRNFLKSVRSLHLKTILLRKTW 330
N++R W +W+++ +L K N+ +S R K + LR TW
Sbjct: 193 NWRRSWGKWKLKWKL*FKRMLNWRRS*RK--WKQVKLR*TW 309
>TC216034
Length = 571
Score = 28.1 bits (61), Expect = 6.0
Identities = 21/79 (26%), Positives = 35/79 (43%), Gaps = 7/79 (8%)
Frame = +3
Query: 124 HLKNRRALLQIQAPKQERVERQDQVQAMMAFHIVVTVVQRVHLM-------LAMKTNRNQ 176
H L + Q +Q+R+E Q ++ + +V VVQ HL+ L T
Sbjct: 138 HTSQHTLLSRYQDQQQQRLESPLQPPGLIMYLLVSLVVQHRHLLPLPLFPQLFYLTVMES 317
Query: 177 LETRREALTSCLLMEPTRR 195
L R+ T+ L +P++R
Sbjct: 318 LRILRKMATAYLSTKPSKR 374
>TC205640 similar to UP|Q9FHW0 (Q9FHW0) Gb|AAF07835.1 (At5g37070), complete
Length = 996
Score = 28.1 bits (61), Expect = 6.0
Identities = 23/101 (22%), Positives = 44/101 (42%), Gaps = 5/101 (4%)
Frame = -3
Query: 132 LQIQAPKQERVERQDQVQAMMAFHIVVTVVQRVHLMLAMKTNRNQLETRREALTSCLLME 191
+++ PKQ++ E + FH +V K+NR + TRR+ L ++
Sbjct: 754 IELNKPKQKKTETTSSTRTQEEFH-------QVSGFTTFKSNRIKWSTRRDNLFY*IVKL 596
Query: 192 PTRRTILLDPFLNHLFLEILLSRYLQLILI-----LEWTYG 227
+R+ LL L++ Y L L+ ++W++G
Sbjct: 595 LNKRSSLLIY*NLSTVTPSLVTSYASLFLVFFMPAVKWSFG 473
>TC205030 UP|ACCD_SOYBN (P49158) Acetyl-coenzyme A carboxylase carboxyl
transferase subunit beta (ACCASE beta chain) , complete
Length = 4503
Score = 28.1 bits (61), Expect = 6.0
Identities = 10/40 (25%), Positives = 24/40 (60%), Gaps = 1/40 (2%)
Frame = -1
Query: 284 SRPSVKNYKRG-WRRWEVRIELSEKSFRNFLKSVRSLHLK 322
+R S+K+++ W +W++ +++ NFL++ +H K
Sbjct: 4146 TRDSIKSHRGPLWNKWKINKVRNKRKITNFLRNKEQIHTK 4027
>TC205936 similar to GB|AAL58101.1|18056655|AF395058 CSN complex subunit 2
{Arabidopsis thaliana;} , partial (56%)
Length = 952
Score = 27.7 bits (60), Expect = 7.8
Identities = 19/56 (33%), Positives = 31/56 (54%)
Frame = +2
Query: 167 MLAMKTNRNQLETRREALTSCLLMEPTRRTILLDPFLNHLFLEILLSRYLQLILIL 222
+LAM TN R E L +++ RRTI+ DPF+ + ++L + Q++L L
Sbjct: 353 ILAM-TNLIAAYQRNEILEFEKILKSNRRTIMDDPFIRNYIEDLLKNIRTQVLLKL 517
>TC205037 similar to UP|Q94II3 (Q94II3) ERD3 protein, partial (83%)
Length = 2018
Score = 27.7 bits (60), Expect = 7.8
Identities = 15/47 (31%), Positives = 21/47 (43%)
Frame = +3
Query: 276 QPGGQDYASRPSVKNYKRGWRRWEVRIELSEKSFRNFLKSVRSLHLK 322
+PGG S P V NY+ WR W IE + + + S+ K
Sbjct: 645 RPGGFWVLSGPPV-NYEHRWRGWNTTIEDQRSDYEKLQELLTSMCFK 782
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.335 0.144 0.454
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,447,632
Number of Sequences: 63676
Number of extensions: 284481
Number of successful extensions: 2678
Number of sequences better than 10.0: 37
Number of HSP's better than 10.0 without gapping: 2639
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2667
length of query: 341
length of database: 12,639,632
effective HSP length: 98
effective length of query: 243
effective length of database: 6,399,384
effective search space: 1555050312
effective search space used: 1555050312
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC148484.3


