

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148482.6 - phase: 0
(402 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
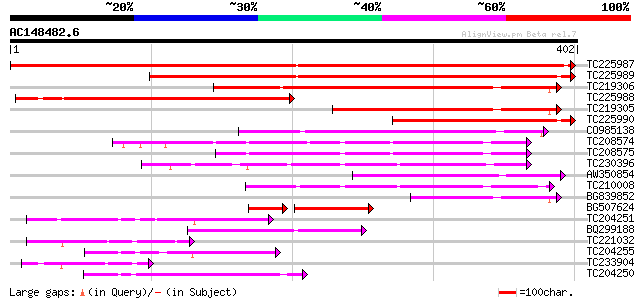
Score E
Sequences producing significant alignments: (bits) Value
TC225987 weakly similar to UP|Q8VYH6 (Q8VYH6) AT4g17280/dl4675c,... 592 e-169
TC225989 weakly similar to UP|Q8VYH6 (Q8VYH6) AT4g17280/dl4675c,... 431 e-121
TC219306 weakly similar to UP|Q9LSE7 (Q9LSE7) Emb|CAB45497.1 (AT... 259 2e-69
TC225988 weakly similar to UP|Q8VYH6 (Q8VYH6) AT4g17280/dl4675c,... 237 6e-63
TC219305 weakly similar to UP|Q9LSE7 (Q9LSE7) Emb|CAB45497.1 (AT... 196 1e-50
TC225990 190 1e-48
CO985138 168 5e-42
TC208574 weakly similar to UP|Q9SSF3 (Q9SSF3) MLP3.2 protein, pa... 145 2e-35
TC208575 weakly similar to UP|Q9SSF3 (Q9SSF3) MLP3.2 protein, pa... 138 5e-33
TC230396 122 4e-28
AW350854 similar to GP|20197988|gb expressed protein {Arabidopsi... 117 7e-27
TC210008 114 6e-26
BG839852 97 1e-20
BG507624 83 9e-20
TC204251 weakly similar to UP|O36027 (O36027) Wiskott-Aldrich Sy... 82 6e-16
BQ299188 80 1e-15
TC221032 similar to UP|GP1_CHLRE (Q9FPQ6) Vegetative cell wall p... 75 5e-14
TC204255 PDB|1OD5_A.0|33357661|1OD5_A Chain A, Crystal Structure... 65 5e-11
TC233904 64 1e-10
TC204250 similar to UP|O96030 (O96030) Neurotrophic factor artem... 62 4e-10
>TC225987 weakly similar to UP|Q8VYH6 (Q8VYH6) AT4g17280/dl4675c, partial
(45%)
Length = 1703
Score = 592 bits (1526), Expect = e-169
Identities = 289/403 (71%), Positives = 336/403 (82%), Gaps = 2/403 (0%)
Frame = +2
Query: 1 MVKNLRFMFLISILMFVTTTLAQTTTQT-CKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQ 59
MV L + L+ + +++ L T+ QT CK Q FT N +FT+CRDLP L+SYLHWT+DQ
Sbjct: 218 MVGKLVVVRLVLAISVLSSLLLTTSAQTACKGQAFTENKVFTTCRDLPHLSSYLHWTFDQ 397
Query: 60 TTGKLDIAFRHKGITDTNRWVAWAINPNNDLASSMNGAQALVAILQSSGTPKAYTSSIAN 119
TGKLDIAFRH GI+ T++WVAWAINP+N+L S+M GAQALVAI+ SSG P AYTSSIAN
Sbjct: 398 ATGKLDIAFRHTGISGTDKWVAWAINPSNNLNSAMTGAQALVAIIPSSGAPNAYTSSIAN 577
Query: 120 SRTQLAESNISYPHSGLIATHENNEVTIYASITLPVGTPSLVHLWQDGAMSGSTPQMHDM 179
T LAE ISY HSGL ATH++ EVTIYA++TLP GT +LVHLW DG +S TP MH M
Sbjct: 578 PGTTLAEGAISYNHSGLTATHQSTEVTIYATLTLPSGTTTLVHLWNDGPVSSGTPAMHSM 757
Query: 180 TSANTQSKESLDLRSGASEQGSGGGSLSRRRNTHGVLNAISWGILMPLGAVIARYLKVFK 239
TS+NTQSKESLDL SG+S+ GSG SL RRRN HGVLNA+SWGILMP+GA+IARYLKVFK
Sbjct: 758 TSSNTQSKESLDLLSGSSQAGSGN-SLRRRRNVHGVLNALSWGILMPVGAIIARYLKVFK 934
Query: 240 SADPAWFYLHVTCQSAAYIVGVAGWGTGLKLGSDSAGVTYSTHRTLGIVIFCLGTLQVFA 299
SADPAWFYLHVTCQ++AYIVGVAGWGTGLKLGSDS G+ Y+THR LGI +FCLGTLQVFA
Sbjct: 935 SADPAWFYLHVTCQTSAYIVGVAGWGTGLKLGSDSVGIKYNTHRALGITLFCLGTLQVFA 1114
Query: 300 LLLRPKKDHKIRFYWNLYHWGVGYATIIISIINIFKGFEALEVSAADRYDNWKHAYTGII 359
LLLRP KDHKIR YWN+YH+ VGY+TIIISIIN+FKGF+ALE S DRY++WKHAY GII
Sbjct: 1115LLLRPNKDHKIRIYWNIYHYAVGYSTIIISIINVFKGFDALETSVGDRYNDWKHAYIGII 1294
Query: 360 AALGGVAVLLEAYTWIIVIKRKKSENK-LQGMNGTNGNGYGSR 401
AALGG+AVLLEAYTWI+V+KR+ SENK G+NGT NGY SR
Sbjct: 1295AALGGIAVLLEAYTWIVVLKRRNSENKTAHGVNGT--NGYDSR 1417
>TC225989 weakly similar to UP|Q8VYH6 (Q8VYH6) AT4g17280/dl4675c, partial
(41%)
Length = 1233
Score = 431 bits (1107), Expect = e-121
Identities = 210/303 (69%), Positives = 252/303 (82%), Gaps = 1/303 (0%)
Frame = +3
Query: 100 LVAILQSSGTPKAYTSSIANSRTQLAESNISYPHSGLIATHENNEVTIYASITLPVGTPS 159
LVAI QS+G+P+AYTSSIA++ T L E ISYP SGL AT E+N+VTI+A++TLP GT S
Sbjct: 3 LVAIPQSNGSPRAYTSSIASTSTTLEEGAISYPVSGLSATFESNQVTIFATLTLPNGTSS 182
Query: 160 LVHLWQDGAMSGSTPQMHDMTSANTQSKESLDLRSGASEQGSGGGSLSRRRNTHGVLNAI 219
LVH+WQDG +SG+TPQ H +++ SKE LDL SG+S Q +G S +RRNTHGVLNA+
Sbjct: 183 LVHVWQDGPLSGTTPQEHSHETSHQNSKEILDLLSGSSTQATGN-SRQKRRNTHGVLNAV 359
Query: 220 SWGILMPLGAVIARYLKVFKSADPAWFYLHVTCQSAAYIVGVAGWGTGLKLGSDSAGVTY 279
SWGILMP GA+IARYLKVFKSADPAWFYLH+TCQ++AYIVGV+G+GTGLKLGSDS GV Y
Sbjct: 360 SWGILMPTGAIIARYLKVFKSADPAWFYLHITCQASAYIVGVSGFGTGLKLGSDSEGVEY 539
Query: 280 STHRTLGIVIFCLGTLQVFALLLRPKKDHKIRFYWNLYHWGVGYATIIISIINIFKGFEA 339
THR LGIV+ CLGTLQVFAL LRP KDH+ R YWN+YH VGYATIIIS++N+FKGF+
Sbjct: 540 DTHRALGIVLVCLGTLQVFALFLRPNKDHRYRVYWNVYHHLVGYATIIISVVNVFKGFDT 719
Query: 340 LEVSAADRYDNWKHAYTGIIAALGGVAVLLEAYTWIIVIKRKKSENKLQGMNGTNG-NGY 398
+E+ DRY++WKHAY GII ALGG+AV LEA+TWIIV KR+KSENK+ +G NG NGY
Sbjct: 720 IEIYVGDRYNSWKHAYIGIIGALGGIAVFLEAFTWIIVFKRRKSENKIP--HGANGVNGY 893
Query: 399 GSR 401
GSR
Sbjct: 894 GSR 902
>TC219306 weakly similar to UP|Q9LSE7 (Q9LSE7) Emb|CAB45497.1
(AT3g25290/MJL12_25), partial (46%)
Length = 939
Score = 259 bits (661), Expect = 2e-69
Identities = 122/250 (48%), Positives = 165/250 (65%), Gaps = 3/250 (1%)
Frame = +1
Query: 145 VTIYASITLPVGTPSLVHLWQDG-AMSGSTPQMHDMTSANTQSKESLDLRSGASEQGSGG 203
+ I+A++ +P S+ H+WQ G +++ HD +N SK L +GA G
Sbjct: 22 IRIFATVKVPEKVESVNHVWQVGPSVTAGRIDRHDFGPSNMNSKGVLSF-NGAQVGGGAV 198
Query: 204 GSLSRRRNTHGVLNAISWGILMPLGAVIARYLKVFKSADPAWFYLHVTCQSAAYIVGVAG 263
++ ++N HG+LNA+SWG+L PLG ++ARY++ F SADPAWFYLHV CQ +AY +GVAG
Sbjct: 199 DPITIKKNIHGILNAVSWGVLFPLGVIVARYMRTFPSADPAWFYLHVGCQVSAYAIGVAG 378
Query: 264 WGTGLKLGSDSAGVTYSTHRTLGIVIFCLGTLQVFALLLRPKKDHKIRFYWNLYHWGVGY 323
WGTG+KLGS+S G+ Y +HR +GI +FC TLQ+FAL LRP KDHK R+ WN+YH VGY
Sbjct: 379 WGTGMKLGSESVGIQYRSHRYIGIALFCFATLQIFALFLRPVKDHKYRYIWNIYHHSVGY 558
Query: 324 ATIIISIINIFKGFEALEVSAADRYDNWKHAYTGIIAALGGVAVLLEAYTWIIVIKRK-- 381
+ +I+ IINIF+GF L WK YT ++ ALG VA+ LE TWI+V+KRK
Sbjct: 559 SIVILGIINIFRGFSILHPD-----QKWKSTYTAVLIALGAVALFLEVITWIVVLKRKSY 723
Query: 382 KSENKLQGMN 391
KS G N
Sbjct: 724 KSTKTYDGYN 753
>TC225988 weakly similar to UP|Q8VYH6 (Q8VYH6) AT4g17280/dl4675c, partial
(12%)
Length = 752
Score = 237 bits (605), Expect = 6e-63
Identities = 118/198 (59%), Positives = 151/198 (75%)
Frame = +1
Query: 5 LRFMFLISILMFVTTTLAQTTTQTCKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQTTGKL 64
LR + +S+L V T ++ QTC++Q F+N +F +CRDLPQLT+YLHWTYDQ +G+L
Sbjct: 163 LRVVLGLSVLSCVLVT---SSAQTCRNQTFSNR-VFATCRDLPQLTAYLHWTYDQASGRL 330
Query: 65 DIAFRHKGITDTNRWVAWAINPNNDLASSMNGAQALVAILQSSGTPKAYTSSIANSRTQL 124
DIAF+H GIT TNRWVAWAINP N L +M GAQALVAI QS+G+P+AYTSSI ++ T L
Sbjct: 331 DIAFKHAGITSTNRWVAWAINPRNTLDPAMIGAQALVAIPQSNGSPRAYTSSITSTSTTL 510
Query: 125 AESNISYPHSGLIATHENNEVTIYASITLPVGTPSLVHLWQDGAMSGSTPQMHDMTSANT 184
E ISYP SGL AT ENN+VTI+A++TLP GT S VH+WQDG +SG+TP+ H +++
Sbjct: 511 EEGAISYPLSGLSATFENNQVTIFATLTLPNGTTSFVHVWQDGPLSGTTPREHSHETSHQ 690
Query: 185 QSKESLDLRSGASEQGSG 202
SKE LDL SG+S Q +G
Sbjct: 691 NSKEILDLLSGSSTQPTG 744
>TC219305 weakly similar to UP|Q9LSE7 (Q9LSE7) Emb|CAB45497.1
(AT3g25290/MJL12_25), partial (39%)
Length = 627
Score = 196 bits (499), Expect = 1e-50
Identities = 91/164 (55%), Positives = 113/164 (68%), Gaps = 2/164 (1%)
Frame = +3
Query: 230 VIARYLKVFKSADPAWFYLHVTCQSAAYIVGVAGWGTGLKLGSDSAGVTYSTHRTLGIVI 289
+IARY++ F ADPAWFYLHV CQ ++Y +GVAGWGTG+KLGS S G+ YS HR +GI +
Sbjct: 6 IIARYMRTFXXADPAWFYLHVGCQVSSYAIGVAGWGTGMKLGSQSEGIQYSAHRYIGIFL 185
Query: 290 FCLGTLQVFALLLRPKKDHKIRFYWNLYHWGVGYATIIISIINIFKGFEALEVSAADRYD 349
F TLQ+FAL LRP KDHK R+ WN+YH +GY+ II+ IINIF+GF L
Sbjct: 186 FSFATLQIFALFLRPVKDHKYRYIWNIYHHSIGYSIIILGIINIFRGFSILHPD-----Q 350
Query: 350 NWKHAYTGIIAALGGVAVLLEAYTWIIVIKRK--KSENKLQGMN 391
WK YT ++ ALG VA+ LE TWI+V+KRK KS G N
Sbjct: 351 KWKSTYTAVLIALGAVALFLEVITWIVVLKRKSGKSTKTYDGHN 482
>TC225990
Length = 669
Score = 190 bits (482), Expect = 1e-48
Identities = 92/131 (70%), Positives = 105/131 (79%), Gaps = 1/131 (0%)
Frame = +1
Query: 272 SDSAGVTYSTHRTLGIVIFCLGTLQVFALLLRPKKDHKIRFYWNLYHWGVGYATIIISII 331
SDS GV Y THR LGIV+ CLGTLQVFAL LRP KDHK R YWN+YH VGYATIIIS++
Sbjct: 1 SDSEGVDYDTHRALGIVLVCLGTLQVFALFLRPNKDHKYRVYWNVYHHLVGYATIIISVV 180
Query: 332 NIFKGFEALEVSAADRYDNWKHAYTGIIAALGGVAVLLEAYTWIIVIKRKKSENKLQGMN 391
NIF+GFE +E DRY++WKHAY GII AL G+AV LEA+TWIIV KR+KSENK+ +
Sbjct: 181 NIFEGFETIEKYVGDRYNSWKHAYIGIIGALAGIAVFLEAFTWIIVFKRRKSENKIP--H 354
Query: 392 GTNG-NGYGSR 401
G NG NGYGSR
Sbjct: 355 GANGANGYGSR 387
>CO985138
Length = 847
Score = 168 bits (425), Expect = 5e-42
Identities = 92/224 (41%), Positives = 119/224 (53%), Gaps = 4/224 (1%)
Frame = -3
Query: 163 LWQDGA-MSGSTPQMHDMTSANTQSKESLDLRSGASEQGSGGGSLSRRRNTHGVLNAISW 221
+WQ G + P H N S E +DL G L R+ HGVLN I W
Sbjct: 845 VWQVGYDIEDGHPLGHPTXXRNXDSTEVIDLTDNGRSTGQYRSYL---RSVHGVLNIIGW 675
Query: 222 GILMPLGAVIARYLKVFKSA-DPAWFYLHVTCQSAAYIVGVAGWGTGLKLGSDSAGVTYS 280
G L+P+G + ARY +VF +P WF LH+ CQ ++VG+ GW GL LG S T+
Sbjct: 674 GTLLPIGIITARYFRVFPFKWEPMWFNLHIGCQLTGFLVGITGWAIGLSLGHSSRYYTFH 495
Query: 281 THRTLGIVIFCLGTLQVFALLLRPKKDHKIRFYWNLYHWGVGYATIIISIINIFKGFEAL 340
HR GI+IF L T+Q+ A L+PK R YWN+YH +GY + I INIFKG L
Sbjct: 494 AHRNYGILIFTLSTVQMLAFRLKPKVTDDYRKYWNMYHHFLGYGLLAIIFINIFKGITIL 315
Query: 341 EVSAADRYDNWKHAYTGIIAALGGVAVLLEAYTWI--IVIKRKK 382
E A WK Y G +A LG +A LE +TWI ++K K+
Sbjct: 314 EGGVA-----WKWGYIGNLALLGTIAFGLEVFTWIRFFMLKHKQ 198
>TC208574 weakly similar to UP|Q9SSF3 (Q9SSF3) MLP3.2 protein, partial (31%)
Length = 1269
Score = 145 bits (367), Expect = 2e-35
Identities = 97/309 (31%), Positives = 151/309 (48%), Gaps = 12/309 (3%)
Frame = +1
Query: 74 TDTNRW--VAWAINPNNDLA------SSMNGAQALVAILQSSGT----PKAYTSSIANSR 121
T N W + A NPN+ +A M G+ A+V + S+G + Y + + ++
Sbjct: 154 TSQNIWSFILSAPNPNSYIAIGFSPNGGMVGSSAIVGWISSNGAGGGMKQYYLTGLTPNQ 333
Query: 122 TQLAESNISYPHSGLIATHENNEVTIYASITLPVGTPSLVHLWQDGAMSGSTPQMHDMTS 181
N+ + T +++ +Y + L P ++ G +G P
Sbjct: 334 VVPDRGNLKVLTNSTFITSQSSR--LYMAFQLETNQPLSKLIYAFGP-NGVFPSAPSFAL 504
Query: 182 ANTQSKESLDLRSGASEQGSGGGSLSRRRNTHGVLNAISWGILMPLGAVIARYLKVFKSA 241
A Q K S+ L + G S + +R+ HG+LN + WGIL+ +GA++ARY FK
Sbjct: 505 AQHQDKVSITLNYATGSSATTGKSYNLKRS-HGLLNILGWGILIIMGAIVARY---FKEW 672
Query: 242 DPAWFYLHVTCQSAAYIVGVAGWGTGLKLGSDSAGVTYSTHRTLGIVIFCLGTLQVFALL 301
DP WFY H + QS +++G+ G +G L ++ S H+ LGI+IF LG LQ+ ALL
Sbjct: 673 DPFWFYFHASVQSLGFVLGIVGVISGFVL-NNQLHTDVSLHKALGIIIFVLGCLQIMALL 849
Query: 302 LRPKKDHKIRFYWNLYHWGVGYATIIISIINIFKGFEALEVSAADRYDNWKHAYTGIIAA 361
RPKK+ K+R YWN YH +G II++I NIF G + W Y ++A
Sbjct: 850 GRPKKESKVRKYWNAYHHNMGRILIILAIANIFYG-----IKLGKEGSGWNIGYGIVLAV 1014
Query: 362 LGGVAVLLE 370
L +A+ E
Sbjct: 1015LFTMAITFE 1041
>TC208575 weakly similar to UP|Q9SSF3 (Q9SSF3) MLP3.2 protein, partial (17%)
Length = 907
Score = 138 bits (347), Expect = 5e-33
Identities = 79/224 (35%), Positives = 115/224 (51%)
Frame = +1
Query: 147 IYASITLPVGTPSLVHLWQDGAMSGSTPQMHDMTSANTQSKESLDLRSGASEQGSGGGSL 206
+Y + L P ++ G +G P + Q K S+ L + G S
Sbjct: 55 LYMAFQLQTNQPLSKLIYAFGP-NGVFPSAPTFSLTQHQDKVSITLNYSTGSSATTGNSY 231
Query: 207 SRRRNTHGVLNAISWGILMPLGAVIARYLKVFKSADPAWFYLHVTCQSAAYIVGVAGWGT 266
+ + +HG+LN WGIL+ +GA++ARY FK DP WFY H + QS +++GV G +
Sbjct: 232 TILKRSHGILNIFGWGILIIMGAIVARY---FKEWDPFWFYFHASVQSLGFVLGVTGVIS 402
Query: 267 GLKLGSDSAGVTYSTHRTLGIVIFCLGTLQVFALLLRPKKDHKIRFYWNLYHWGVGYATI 326
G L ++ S H+ LGI+IF L LQ+ ALL RPKK+ K+R YWNLYH +G I
Sbjct: 403 GFVL-NNQLHTDVSLHKVLGIIIFVLACLQIMALLGRPKKESKVRKYWNLYHHNLGRILI 579
Query: 327 IISIINIFKGFEALEVSAADRYDNWKHAYTGIIAALGGVAVLLE 370
I++I NIF G + W Y ++A L +A+ E
Sbjct: 580 ILAIANIFYG-----IKLGKEGSGWNIGYGIVLAVLLTMAITFE 696
>TC230396
Length = 1001
Score = 122 bits (305), Expect = 4e-28
Identities = 88/284 (30%), Positives = 130/284 (44%), Gaps = 7/284 (2%)
Frame = +3
Query: 94 MNGAQALVAILQSSGTPKA---YTSSIANSRTQLAESNISYPHSGLIATHENNEVTIYAS 150
M G+ A+V + G K Y S ++ + + P + + A N I+ +
Sbjct: 195 MVGSSAMVGWITKHGHAKIKQFYLRGRKQSEVKIDKGEL--PLNNIPAAVATNGAEIHIA 368
Query: 151 ITLPVGTPSLVHLWQDG----AMSGSTPQMHDMTSANTQSKESLDLRSGASEQGSGGGSL 206
L + TP +Q A PQ H ++ ++ D +G++ G L
Sbjct: 369 FQLQMTTP-----FQKQPILLAFGSKYPQNHHLSKHEDKTAIVFDFSAGST--GPVSREL 527
Query: 207 SRRRNTHGVLNAISWGILMPLGAVIARYLKVFKSADPAWFYLHVTCQSAAYIVGVAGWGT 266
+ R HG+L I WG+++P+GA+IARY F+ DP WFYLH Q + G+
Sbjct: 528 IQMRTNHGILAIIGWGLILPVGAIIARY---FRHKDPLWFYLHAIIQFVGFTFGLGTVIL 698
Query: 267 GLKLGSDSAGVTYSTHRTLGIVIFCLGTLQVFALLLRPKKDHKIRFYWNLYHWGVGYATI 326
GL+L S V HR +GI L LQV AL LRP KD KIR +WN YH G +
Sbjct: 699 GLQLYSKMQ-VHIPAHRGIGIFALVLSILQVLALFLRPNKDSKIRKFWNWYHRWFGRMAL 875
Query: 327 IISIINIFKGFEALEVSAADRYDNWKHAYTGIIAALGGVAVLLE 370
+ + INI G + AA +WK Y + + A++LE
Sbjct: 876 VFAAINIVLGMQ-----AAGAGSDWKIGYGFVFGIMVVAAIVLE 992
>AW350854 similar to GP|20197988|gb expressed protein {Arabidopsis thaliana},
partial (35%)
Length = 538
Score = 117 bits (294), Expect = 7e-27
Identities = 56/152 (36%), Positives = 88/152 (57%), Gaps = 1/152 (0%)
Frame = -1
Query: 244 AWFYLHVTCQSAAYIVGVAGWGTGLKLGSDSAGVTYSTHRTLGIVIFCLGTLQVFALLLR 303
AWFY H Q +++G G+ G++LG S GV Y HR LG+ +FCLG LQ ALL R
Sbjct: 538 AWFYAHAGMQVFGFVLGTVGFVIGIRLGQLSPGVEYRLHRKLGMAVFCLGGLQTLALLFR 359
Query: 304 PKKDHKIRFYWNLYHWGVGYATIIISIINIFKGFEALEVSAADRYDNWKHAYTGIIAALG 363
P +K R YW YH VGY+ +++ +N+F+GFE + S + K Y ++ L
Sbjct: 358 PNTRNKFRKYWKSYHHFVGYSCVVLGFVNVFQGFEVMGASRSYA----KLTYCLGLSTLI 191
Query: 364 GVAVLLEAYTWIIVIKRKKSEN-KLQGMNGTN 394
G+ + LE +W++ ++ K + + +G+ GT+
Sbjct: 190 GLCIALEVNSWVVFCRKSKEDKMRREGLIGTS 95
>TC210008
Length = 1054
Score = 114 bits (286), Expect = 6e-26
Identities = 71/219 (32%), Positives = 109/219 (49%)
Frame = +2
Query: 168 AMSGSTPQMHDMTSANTQSKESLDLRSGASEQGSGGGSLSRRRNTHGVLNAISWGILMPL 227
A S P H ++ ++ D SG++ S G L R +HG++ I WG+++P+
Sbjct: 146 AFSTKHPLNHHLSKHVDKAAIIFDFSSGSTGPVSNG--LIHMRKSHGIVGIIGWGLILPV 319
Query: 228 GAVIARYLKVFKSADPAWFYLHVTCQSAAYIVGVAGWGTGLKLGSDSAGVTYSTHRTLGI 287
GA+IARY F+ DP WFYLH Q + G+ GL+L + V HR +GI
Sbjct: 320 GAIIARY---FRHKDPLWFYLHSVIQFVGFSFGLGTVLLGLQLYRNMH-VHIPAHRGIGI 487
Query: 288 VIFCLGTLQVFALLLRPKKDHKIRFYWNLYHWGVGYATIIISIINIFKGFEALEVSAADR 347
+ L LQ+ A LRP KD K R WNLYH G + + +NI G + AA
Sbjct: 488 FVLVLSILQILAFFLRPDKDSKYRNIWNLYHGWFGRMALFFAALNIVLG-----MRAAGA 652
Query: 348 YDNWKHAYTGIIAALGGVAVLLEAYTWIIVIKRKKSENK 386
++WK Y +++ + ++LE ++ K+SE +
Sbjct: 653 GNDWKAGYGFLLSIVLVAVIVLEVLAYL-----KRSEKR 754
>BG839852
Length = 685
Score = 97.1 bits (240), Expect = 1e-20
Identities = 50/109 (45%), Positives = 64/109 (57%), Gaps = 2/109 (1%)
Frame = +2
Query: 285 LGIVIFCLGTLQVFALLLRPKKDHKIRFYWNLYHWGVGYATIIISIINIFKGFEALEVSA 344
L +V + L +FAL LRP KD K R+ WN+YH VGY+ +I+ IIN F+GF L
Sbjct: 191 LAVVSLHVLIL*IFALFLRPVKDXKYRYXWNIYHHSVGYSIVILGIINXFRGFSILHPD- 367
Query: 345 ADRYDNWKHAYTGIIAALGGVAVLLEAYTWIIVIKRK--KSENKLQGMN 391
WK YT ++ ALG VA+ LE TWI+V+KRK KS G N
Sbjct: 368 ----QKWKSTYTAVLIALGAVALFLEVITWIVVLKRKXYKSXKTYDGYN 502
>BG507624
Length = 268
Score = 83.2 bits (204), Expect(2) = 9e-20
Identities = 36/56 (64%), Positives = 46/56 (81%)
Frame = +2
Query: 203 GGSLSRRRNTHGVLNAISWGILMPLGAVIARYLKVFKSADPAWFYLHVTCQSAAYI 258
G S +RRNTHGVLNA+SWG+L+P A+ AR L VFKSA PAWFYLH+T Q++A++
Sbjct: 98 GNSRQKRRNTHGVLNAVSWGLLLPTXALFARSLXVFKSAXPAWFYLHLTXQASAFL 265
Score = 31.6 bits (70), Expect(2) = 9e-20
Identities = 15/28 (53%), Positives = 20/28 (70%)
Frame = +1
Query: 170 SGSTPQMHDMTSANTQSKESLDLRSGAS 197
SG+TPQ H +++ SKE LDL SG+S
Sbjct: 1 SGTTPQEHRHETSHQNSKEILDLLSGSS 84
>TC204251 weakly similar to UP|O36027 (O36027) Wiskott-Aldrich Syndrome
protein homolog, partial (8%)
Length = 1045
Score = 81.6 bits (200), Expect = 6e-16
Identities = 54/178 (30%), Positives = 85/178 (47%), Gaps = 3/178 (1%)
Frame = +2
Query: 13 ILMFVTTTLAQTTTQTCKSQNFTNNAIFTSCRDLPQLTSYLHWTYDQTTGKLDIAFRHKG 72
I++F + + TC SQ N + +C +LP L + LH+T++ T L +AF +
Sbjct: 128 IILFSLFSTPSHSALTCASQKL--NRTYANCTNLPTLGATLHFTFNATNRSLSVAFSAEP 301
Query: 73 ITDTNRWVAWAINPNNDLASSMNGAQALVAILQSSGTPKAYTSSIANSRTQLAESNI--- 129
+ + WVAW +N D M GA+A +A SS + A T N + A +
Sbjct: 302 PSRSG-WVAWGLNLVGD---GMRGAEAFLAF-PSSASASAITLGRYNLTSYKAIDEVKAF 466
Query: 130 SYPHSGLIATHENNEVTIYASITLPVGTPSLVHLWQDGAMSGSTPQMHDMTSANTQSK 187
++ L A N VTIY S+ +P ++ H+WQ G ++ P +H NT SK
Sbjct: 467 TFDSWDLAAEESNGAVTIYGSVKIPDSARNVSHVWQVGPVAAGKPGVHSFEKKNTDSK 640
>BQ299188
Length = 422
Score = 80.5 bits (197), Expect = 1e-15
Identities = 45/129 (34%), Positives = 70/129 (53%), Gaps = 2/129 (1%)
Frame = +1
Query: 127 SNISYPHSGLIAT-HENNEVTIYASITLPVGTPSLVHLWQDGA-MSGSTPQMHDMTSANT 184
S+ + + G +AT H + I+ ++ L + +W G + G +P +H TS +
Sbjct: 37 SSTAAMYGGKMATVHNGAAIQIFGTVKLQTNKTKIHLVWNRGLYVQGYSPTIHPTTSTDL 216
Query: 185 QSKESLDLRSGASEQGSGGGSLSRRRNTHGVLNAISWGILMPLGAVIARYLKVFKSADPA 244
S + D+ SG+S L+ R HG +NAISWGIL+P+GA+ ARYL+ ++ PA
Sbjct: 217 ASIATFDVLSGSSAPQHT--DLTTLRVIHGTVNAISWGILLPMGAITARYLRHIQALGPA 390
Query: 245 WFYLHVTCQ 253
WFY H Q
Sbjct: 391 WFYAHAGIQ 417
>TC221032 similar to UP|GP1_CHLRE (Q9FPQ6) Vegetative cell wall protein gp1
precursor (Hydroxyproline-rich glycoprotein 1), partial
(6%)
Length = 503
Score = 75.1 bits (183), Expect = 5e-14
Identities = 42/121 (34%), Positives = 64/121 (52%), Gaps = 2/121 (1%)
Frame = +1
Query: 13 ILMFVTTTLAQTTTQTCKSQNFTN--NAIFTSCRDLPQLTSYLHWTYDQTTGKLDIAFRH 70
+L+F+T A ++ TC +Q T+ N ++++C DLP L S+LHWT+D T L +AF
Sbjct: 130 LLLFLTLVAATASSLTCTTQKLTDSKNNLYSNCLDLPALDSFLHWTHDPTNASLSVAFA- 306
Query: 71 KGITDTNRWVAWAINPNNDLASSMNGAQALVAILQSSGTPKAYTSSIANSRTQLAESNIS 130
++ WV+W INP A M GAQ L A +G T + S T + +S
Sbjct: 307 AAPPNSGGWVSWGINPT---AIGMQGAQVLAAYKADNGAVTVKTLDL-KSYTAIVPGKLS 474
Query: 131 Y 131
+
Sbjct: 475 F 477
>TC204255 PDB|1OD5_A.0|33357661|1OD5_A Chain A, Crystal Structure Of Glycinin
A3b4 Subunit Homohexamer. {Glycine max;} , partial (6%)
Length = 519
Score = 65.1 bits (157), Expect = 5e-11
Identities = 43/146 (29%), Positives = 69/146 (46%), Gaps = 7/146 (4%)
Frame = +3
Query: 54 HWTYDQTTGKLDIAFRHKGITDTNRWVAWAINPNNDLASSMNGAQALVAILQSSGTPKAY 113
++T++ T L +AF + + + WVAW +N D M GA+A +A S AY
Sbjct: 81 NFTFNATNRSLSVAFSAEPPSRSG-WVAWGLNLVGD---GMRGAEAFLAFPSS-----AY 233
Query: 114 TSSIANSRTQLAESN-------ISYPHSGLIATHENNEVTIYASITLPVGTPSLVHLWQD 166
S+I R L ++ L A N+ +TIY+SI +P ++ H+WQ
Sbjct: 234 ASAITLGRYNLTSYKAIDEIKAFTFDSWDLAAEESNSAITIYSSIKIPNSARNISHIWQV 413
Query: 167 GAMSGSTPQMHDMTSANTQSKESLDL 192
G M+ P +H+ NT SK + +
Sbjct: 414 GPMATGKPGVHNFEKKNTDSKATFPI 491
>TC233904
Length = 419
Score = 63.9 bits (154), Expect = 1e-10
Identities = 36/96 (37%), Positives = 51/96 (52%), Gaps = 2/96 (2%)
Frame = +2
Query: 9 FLISILMFVTTTLAQTTTQTCKSQNFT--NNAIFTSCRDLPQLTSYLHWTYDQTTGKLDI 66
FL+ + +F T + TC +Q T N +F++C DLP L S+LHWT+D L +
Sbjct: 65 FLLLLALFAATA----SCLTCTTQKLTDSNKKLFSNCLDLPSLDSFLHWTHDPANASLSV 232
Query: 67 AFRHKGITDTNRWVAWAINPNNDLASSMNGAQALVA 102
AF + WV+W INP+ + M GAQ L A
Sbjct: 233 AF-VAAPPNPGGWVSWGINPS---GTGMVGAQVLAA 328
>TC204250 similar to UP|O96030 (O96030) Neurotrophic factor artemin
(Pre-pro-neublastin) (Pre-pro-enovin precursor), partial
(8%)
Length = 891
Score = 62.4 bits (150), Expect = 4e-10
Identities = 41/160 (25%), Positives = 75/160 (46%), Gaps = 1/160 (0%)
Frame = +2
Query: 53 LHWTYDQTTGKLDIAFRHKGITDTNRWVAWAINPNNDLASSMNGAQALVAILQSSGTPKA 112
LH+T++ T L +AF + + WVAW +N M GA+AL+A+ +SG+
Sbjct: 11 LHFTFNATNRTLSVAFSASPPSPSG-WVAWGLNL---AGGGMAGAEALLALPSTSGSAVT 178
Query: 113 YTS-SIANSRTQLAESNISYPHSGLIATHENNEVTIYASITLPVGTPSLVHLWQDGAMSG 171
++ + ++ ++ L A N +TIY ++ +P ++ H+WQ G ++
Sbjct: 179 LRRYNLTSYKSIDVVKAFTFESWDLSAEETNGAITIYGTVKIPDSAENVSHVWQVGPVAA 358
Query: 172 STPQMHDMTSANTQSKESLDLRSGASEQGSGGGSLSRRRN 211
P +H N +K +L + + GS G S + N
Sbjct: 359 GVPAVHGFKDDNIHAKAALPV----ALAGSSGSSAASGEN 466
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.133 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,323,588
Number of Sequences: 63676
Number of extensions: 252002
Number of successful extensions: 1452
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 1410
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1428
length of query: 402
length of database: 12,639,632
effective HSP length: 99
effective length of query: 303
effective length of database: 6,335,708
effective search space: 1919719524
effective search space used: 1919719524
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC148482.6


