

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
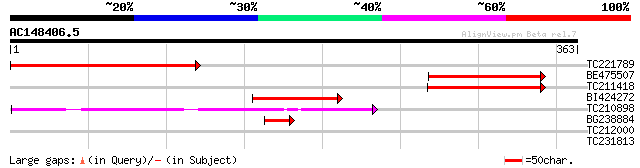
Query= AC148406.5 - phase: 0 /pseudo
(363 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC221789 similar to UP|Q9SXQ6 (Q9SXQ6) FEN-1, partial (37%) 217 6e-57
BE475507 132 2e-31
TC211418 similar to UP|Q9SXQ6 (Q9SXQ6) FEN-1, partial (20%) 128 4e-30
BI424272 homologue to GP|4587225|dbj| FEN-1 {Oryza sativa (japon... 86 3e-17
TC210898 weakly similar to UP|Q8W5R1 (Q8W5R1) Single-strand DNA ... 73 2e-13
BG238884 homologue to PIR|T01198|T011 endonuclease homolog F21E1... 42 3e-04
TC212000 weakly similar to UP|Q892J4 (Q892J4) DNA polymerase I ... 29 3.8
TC231813 28 5.0
>TC221789 similar to UP|Q9SXQ6 (Q9SXQ6) FEN-1, partial (37%)
Length = 520
Score = 217 bits (553), Expect = 6e-57
Identities = 109/122 (89%), Positives = 115/122 (93%)
Frame = +3
Query: 1 MKENKFESYFGRKIAVDASMSIYQFLIVVGRSGTEMLTNEAGEVTSHLQGMFARTIRLLE 60
MKENKFESYFGRKIA+DASMSIYQFLIVVGRSGTEMLTNEAGEVTSHLQGMF+RTIRLLE
Sbjct: 153 MKENKFESYFGRKIAIDASMSIYQFLIVVGRSGTEMLTNEAGEVTSHLQGMFSRTIRLLE 332
Query: 61 AGMKPVYVFDGKPPEMKNQELKKRLSKRAEATAGLTEALEADNKEDIEKFSKRTVKVTKQ 120
AG+KPVYVFDGKPP++K QEL KR SKRAEAT L+EALE NKEDIEKFSKRTVKVTKQ
Sbjct: 333 AGIKPVYVFDGKPPDLKKQELAKRYSKRAEATEDLSEALETANKEDIEKFSKRTVKVTKQ 512
Query: 121 HN 122
HN
Sbjct: 513 HN 518
>BE475507
Length = 374
Score = 132 bits (332), Expect = 2e-31
Identities = 64/75 (85%), Positives = 69/75 (91%)
Frame = +1
Query: 269 EPEVSTDDEVLNLKWSPPDEEGLITFLVNENGFNSDRVTKAIEKIKAAKNKSSQGRLESF 328
EP V TD++ L++KWS PDEEGLITFLVNENGFNSDRVTKAIEKIK AKNKSSQGRLESF
Sbjct: 4 EPMVITDEKELDIKWSSPDEEGLITFLVNENGFNSDRVTKAIEKIKVAKNKSSQGRLESF 183
Query: 329 FKPTANPSVPIKRKQ 343
FKPT NPSVPIKRK+
Sbjct: 184 FKPTVNPSVPIKRKE 228
>TC211418 similar to UP|Q9SXQ6 (Q9SXQ6) FEN-1, partial (20%)
Length = 626
Score = 128 bits (322), Expect = 4e-30
Identities = 64/76 (84%), Positives = 69/76 (90%)
Frame = +1
Query: 268 KEPEVSTDDEVLNLKWSPPDEEGLITFLVNENGFNSDRVTKAIEKIKAAKNKSSQGRLES 327
KEP V TD++ L++K S PDEEGLITFLVNENGFN DRVTKAIEKIK AKNKSSQGRLES
Sbjct: 1 KEPLVITDEKELDIKXSSPDEEGLITFLVNENGFNRDRVTKAIEKIKVAKNKSSQGRLES 180
Query: 328 FFKPTANPSVPIKRKQ 343
FFKPTANPSVPIKRK+
Sbjct: 181 FFKPTANPSVPIKRKE 228
>BI424272 homologue to GP|4587225|dbj| FEN-1 {Oryza sativa (japonica
cultivar-group)}, partial (15%)
Length = 421
Score = 85.9 bits (211), Expect = 3e-17
Identities = 42/58 (72%), Positives = 49/58 (84%)
Frame = +2
Query: 156 VYAVASEDMDSLTFGAPKFLRHLMDPSSKKIPVMEFDVAKILEELDLTMDQFIDLCIL 213
VYAVASEDMDSLTFG+PKFLRHLMDPSSKKIPVMEF+VAK+LE ++ + +C L
Sbjct: 167 VYAVASEDMDSLTFGSPKFLRHLMDPSSKKIPVMEFEVAKVLEAFNILEFFYRMICSL 340
Score = 40.8 bits (94), Expect = 0.001
Identities = 18/21 (85%), Positives = 21/21 (99%)
Frame = +1
Query: 195 KILEELDLTMDQFIDLCILSG 215
+ILEEL++TMDQFIDLCILSG
Sbjct: 358 QILEELNMTMDQFIDLCILSG 420
>TC210898 weakly similar to UP|Q8W5R1 (Q8W5R1) Single-strand DNA
endonuclease-1, partial (6%)
Length = 830
Score = 72.8 bits (177), Expect = 2e-13
Identities = 67/237 (28%), Positives = 106/237 (44%), Gaps = 3/237 (1%)
Frame = +3
Query: 2 KENKFESYFGRKIAVDASMSIYQFLIVVGRSGTEMLTNEAGEVTSHLQGMFARTIRLLEA 61
++ F+ +++AVD S I Q + + HL+ F RTI L
Sbjct: 108 RKEGFDFLRNKRVAVDLSFWIVQHENAIKATHVRK---------PHLRLTFFRTINLFSK 260
Query: 62 -GMKPVYVFDGKPPEMKNQ-ELKKRLSKRAEATAGLTEALEADNKEDIEKFSKRTVKVTK 119
G PV++ DG P +K++ + + A L E + E FS
Sbjct: 261 FGALPVFIVDGTPSPLKSRARIARYFRSSGIELANLPVPEEGVSAERNHMFSS------- 419
Query: 120 QHNDDCKRLLRLMGVPVVEAPSEAEAQCAALCKAGKVYAVASEDMDSLTFGAPKFLRHLM 179
H C L+ L+G+PV++A EAEA CA L G V A + D D+ FGA ++
Sbjct: 420 -HVQKCVELVELLGMPVLKAKGEAEALCAQLNSEGHVDACITADSDAFLFGANCIIK-CF 593
Query: 180 DPSSKKIPVMEFDVAKILEELDLTMDQFIDLCILSGCDY-CDNIRGIGGMTALKLIR 235
P+ K+ P ++++ I L L I + +L G D+ +RGIG TAL+ ++
Sbjct: 594 CPNFKE-PFECYNMSDIEAGLGLKRKHLIAISLLVGNDHDIKGVRGIGLDTALRFVK 761
>BG238884 homologue to PIR|T01198|T011 endonuclease homolog F21E10.3 -
Arabidopsis thaliana, partial (5%)
Length = 113
Score = 42.4 bits (98), Expect = 3e-04
Identities = 18/19 (94%), Positives = 19/19 (99%)
Frame = -1
Query: 164 MDSLTFGAPKFLRHLMDPS 182
MDSLTFG+PKFLRHLMDPS
Sbjct: 59 MDSLTFGSPKFLRHLMDPS 3
>TC212000 weakly similar to UP|Q892J4 (Q892J4) DNA polymerase I , partial
(3%)
Length = 1084
Score = 28.9 bits (63), Expect = 3.8
Identities = 12/23 (52%), Positives = 17/23 (73%)
Frame = +2
Query: 222 IRGIGGMTALKLIRQHGSIEKIL 244
+ G TALKLI++HGS+E +L
Sbjct: 20 VPSFGRKTALKLIKKHGSLETLL 88
>TC231813
Length = 444
Score = 28.5 bits (62), Expect = 5.0
Identities = 16/55 (29%), Positives = 29/55 (52%)
Frame = +2
Query: 70 DGKPPEMKNQELKKRLSKRAEATAGLTEALEADNKEDIEKFSKRTVKVTKQHNDD 124
+ K E++N ++++R+ G T+ALE DN+ D S + ++ Q DD
Sbjct: 29 NSKQVELENIQIQERIMGDDSFNEGKTDALERDNRVD----SMNSSQIEHQQMDD 181
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.135 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,515,769
Number of Sequences: 63676
Number of extensions: 150820
Number of successful extensions: 725
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 722
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 725
length of query: 363
length of database: 12,639,632
effective HSP length: 98
effective length of query: 265
effective length of database: 6,399,384
effective search space: 1695836760
effective search space used: 1695836760
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC148406.5


