

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148404.8 - phase: 0
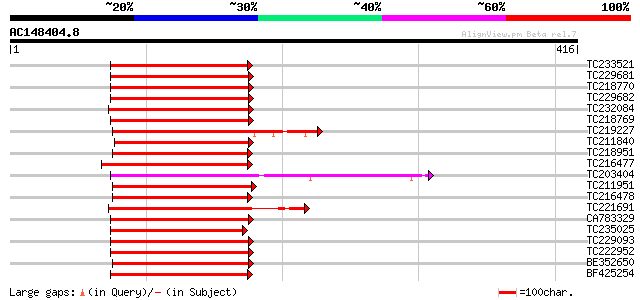
(416 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC233521 similar to UP|O04140 (O04140) Myb protein, partial (44%) 156 2e-38
TC229681 UP|Q9S7E3 (Q9S7E3) GmMYB29A1 protein, complete 152 2e-37
TC218770 UP|Q9XIU5 (Q9XIU5) GmMYB29B2 protein, complete 152 2e-37
TC229682 UP|Q9XIU9 (Q9XIU9) GmMYB29A2 protein, complete 152 2e-37
TC232084 UP|Q84PP2 (Q84PP2) Transcription factor MYB101 (Fragmen... 150 8e-37
TC218769 UP|Q9XIU8 (Q9XIU8) GmMYB29A2 protein, complete 150 1e-36
TC219227 homologue to UP|Q84U52 (Q84U52) MYB2, partial (36%) 146 1e-35
TC211840 homologue to UP|Q8LBF0 (Q8LBF0) Myb-related protein M4,... 145 3e-35
TC218951 homologue to UP|P93474 (P93474) Myb26, partial (77%) 144 6e-35
TC216477 homologue to UP|P93474 (P93474) Myb26, partial (67%) 143 2e-34
TC203404 GmMYB16 142 2e-34
TC211951 similar to GB|AAS10022.1|41619094|AY519552 MYB transcri... 142 2e-34
TC216478 similar to UP|P93474 (P93474) Myb26, partial (83%) 142 4e-34
TC221691 GmMYB6 142 4e-34
CA783329 homologue to GP|6651292|gb| myb-related transcription f... 142 4e-34
TC235025 similar to UP|Q6R077 (Q6R077) MYB transcription factor,... 141 6e-34
TC229093 homologue to PIR|T51621|T51621 myb-like protein [import... 141 6e-34
TC222952 homologue to UP|Q40173 (Q40173) Myb-related transcripti... 141 6e-34
BE352650 140 8e-34
BF425254 homologue to PIR|S26605|S26 myb-related protein 1 - gar... 139 2e-33
>TC233521 similar to UP|O04140 (O04140) Myb protein, partial (44%)
Length = 886
Score = 156 bits (394), Expect = 2e-38
Identities = 66/104 (63%), Positives = 85/104 (81%)
Frame = +2
Query: 75 GLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAI 134
GLKKGPWTA ED+ILV+H+Q+YG GNW ++ K GL RCGKSCRLRW N+LRPD+++G
Sbjct: 50 GLKKGPWTAEEDQILVSHIQRYGHGNWRALPKQAGLLRCGKSCRLRWINYLRPDIKRGKF 229
Query: 135 TAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKK 178
+ EEE I++LH +GN+W+ +AA LPGRTDNEIKNFW+T K+
Sbjct: 230 SKEEEDTILKLHGILGNRWSAIAASLPGRTDNEIKNFWHTHLKR 361
>TC229681 UP|Q9S7E3 (Q9S7E3) GmMYB29A1 protein, complete
Length = 795
Score = 152 bits (385), Expect = 2e-37
Identities = 64/105 (60%), Positives = 85/105 (80%)
Frame = +1
Query: 75 GLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAI 134
GLKKGPWT ED+IL++++QK+G GNW ++ K GL RCGKSCRLRW N+LRPD+++G
Sbjct: 31 GLKKGPWTPEEDQILMSYIQKHGHGNWRALPKLAGLLRCGKSCRLRWINYLRPDIKRGNF 210
Query: 135 TAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
++EEE II++H +GN+W+ +AA LPGRTDNEIKN W+T KKR
Sbjct: 211 SSEEEEIIIKMHELLGNRWSAIAAKLPGRTDNEIKNVWHTHLKKR 345
>TC218770 UP|Q9XIU5 (Q9XIU5) GmMYB29B2 protein, complete
Length = 1316
Score = 152 bits (385), Expect = 2e-37
Identities = 65/105 (61%), Positives = 82/105 (77%)
Frame = +1
Query: 75 GLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAI 134
GLKKGPW ED+IL +++QK+G GNW ++ K GL RCGKSCRLRW N+LRPD+++G
Sbjct: 142 GLKKGPWATEEDQILTSYIQKHGHGNWRALPKQAGLLRCGKSCRLRWINYLRPDIKRGNF 321
Query: 135 TAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
T EEE II+LH +GN+W+ +AA LPGRTDNEIKN W+T KKR
Sbjct: 322 TIEEEETIIKLHEMLGNRWSAIAAKLPGRTDNEIKNVWHTNLKKR 456
>TC229682 UP|Q9XIU9 (Q9XIU9) GmMYB29A2 protein, complete
Length = 1249
Score = 152 bits (385), Expect = 2e-37
Identities = 64/105 (60%), Positives = 85/105 (80%)
Frame = +3
Query: 75 GLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAI 134
GLKKGPWT ED+IL++++QK+G GNW ++ K GL RCGKSCRLRW N+LRPD+++G
Sbjct: 114 GLKKGPWTPEEDQILMSYIQKHGHGNWRALPKLAGLLRCGKSCRLRWINYLRPDIKRGNF 293
Query: 135 TAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
++EEE II++H +GN+W+ +AA LPGRTDNEIKN W+T KKR
Sbjct: 294 SSEEEEIIIKMHELLGNRWSAIAAKLPGRTDNEIKNVWHTHLKKR 428
>TC232084 UP|Q84PP2 (Q84PP2) Transcription factor MYB101 (Fragment), complete
Length = 1383
Score = 150 bits (380), Expect = 8e-37
Identities = 66/107 (61%), Positives = 82/107 (75%)
Frame = +2
Query: 73 EGGLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKG 132
E GLKKGPWT ED ILV ++QK+G G+W ++ K L RCGKSCRLRW N+LRPD+++G
Sbjct: 77 ENGLKKGPWTPEEDRILVDYIQKHGHGSWRALPKHARLNRCGKSCRLRWTNYLRPDIKRG 256
Query: 133 AITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
+ EEE+ II LH +GNKW+ +A LPGRTDNEIKNFWNT KK+
Sbjct: 257 KFSEEEEQLIINLHAVLGNKWSAIAGHLPGRTDNEIKNFWNTHLKKK 397
>TC218769 UP|Q9XIU8 (Q9XIU8) GmMYB29A2 protein, complete
Length = 828
Score = 150 bits (378), Expect = 1e-36
Identities = 64/105 (60%), Positives = 81/105 (76%)
Frame = +1
Query: 75 GLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAI 134
GLKKGPW ED+IL +++ K+G GNW ++ K GL RCGKSCRLRW N+LRPD+++G
Sbjct: 31 GLKKGPWAPEEDQILTSYIDKHGHGNWRALPKQAGLLRCGKSCRLRWINYLRPDIKRGNF 210
Query: 135 TAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
T EEE II+LH +GN+W+ +AA LPGRTDNEIKN W+T KKR
Sbjct: 211 TIEEEETIIKLHDMLGNRWSAIAAKLPGRTDNEIKNVWHTNLKKR 345
>TC219227 homologue to UP|Q84U52 (Q84U52) MYB2, partial (36%)
Length = 1311
Score = 146 bits (369), Expect = 1e-35
Identities = 81/169 (47%), Positives = 106/169 (61%), Gaps = 15/169 (8%)
Frame = +1
Query: 76 LKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAIT 135
L+KG W+ EDE L+ H+ KYG G W+SV K GL RCGKSCRLRW N+LRPDL +G +
Sbjct: 37 LRKGLWSPEEDEKLLRHITKYGHGCWSSVPKQAGLQRCGKSCRLRWINYLRPDLNRGTFS 216
Query: 136 AEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKK--RGRANLPIYPEDLS- 192
EEE IIELH +GN+W+Q+AA LPGRTDNEIKN WN+ KK R R P+ + LS
Sbjct: 217 QEEETLIIELHAVLGNRWSQIAAQLPGRTDNEIKNLWNSCLKKKLRQRGIDPVTHKPLSE 396
Query: 193 ----------SECLLNQENADMLTNEASQHDEA--ENKVPEVIFKDYKL 229
S+ L N+ N +L +E+ + DE E + + K Y++
Sbjct: 397 VENGEEDKTRSQELSNELN--LLNSESFKSDEGSYEQRASSIAPKAYEM 537
>TC211840 homologue to UP|Q8LBF0 (Q8LBF0) Myb-related protein M4, partial
(38%)
Length = 516
Score = 145 bits (367), Expect = 3e-35
Identities = 61/102 (59%), Positives = 80/102 (77%)
Frame = +2
Query: 78 KGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAITAE 137
KGPWT ED+ L+ ++QK+G GNW ++ K GL RCGKSCRLRW N+LRPD+++G + E
Sbjct: 2 KGPWTTEEDQKLIDYIQKHGYGNWRTLPKNAGLQRCGKSCRLRWTNYLRPDIKRGRFSFE 181
Query: 138 EERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
EE II+LH +GNKW+ +A+ LPGRTDNEIKN+WNT +KR
Sbjct: 182 EEETIIQLHSILGNKWSAIASRLPGRTDNEIKNYWNTHIRKR 307
>TC218951 homologue to UP|P93474 (P93474) Myb26, partial (77%)
Length = 866
Score = 144 bits (364), Expect = 6e-35
Identities = 64/103 (62%), Positives = 81/103 (78%)
Frame = +3
Query: 76 LKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAIT 135
++KGPWT ED IL+ ++ +GEG WNS+ K +GL R GKSCRLRW N+LRPD+R+G IT
Sbjct: 48 VRKGPWTMEEDLILINYIANHGEGVWNSLAKASGLKRTGKSCRLRWLNYLRPDVRRGNIT 227
Query: 136 AEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKK 178
EE+ IIELH K GN+W+++A LPGRTDNEIKNFW TR +K
Sbjct: 228 PEEQLLIIELHAKWGNRWSKIAKHLPGRTDNEIKNFWRTRIQK 356
>TC216477 homologue to UP|P93474 (P93474) Myb26, partial (67%)
Length = 1001
Score = 143 bits (360), Expect = 2e-34
Identities = 63/111 (56%), Positives = 82/111 (73%)
Frame = +2
Query: 68 GGEGGEGGLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRP 127
G + ++KGPWT ED IL+ ++ +GEG WNS+ K GL R GKSCRLRW N+LRP
Sbjct: 44 GNTSHDPEVRKGPWTMEEDLILITYIANHGEGVWNSLAKAAGLKRTGKSCRLRWLNYLRP 223
Query: 128 DLRKGAITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKK 178
D+R+G IT EE+ I+ELH K GN+W+++A LPGRTDNEIKN+W TR +K
Sbjct: 224 DVRRGNITPEEQLLIMELHAKWGNRWSKIAKHLPGRTDNEIKNYWRTRIQK 376
>TC203404 GmMYB16
Length = 1138
Score = 142 bits (359), Expect = 2e-34
Identities = 81/242 (33%), Positives = 132/242 (54%), Gaps = 5/242 (2%)
Frame = +1
Query: 75 GLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAI 134
GL KGPWT ED +L ++Q +GEG W S+ K GL RCGKSCRLRW N+LRPD+++G I
Sbjct: 136 GLHKGPWTPKEDALLTKYIQAHGEGQWKSLPKKAGLLRCGKSCRLRWMNYLRPDIKRGNI 315
Query: 135 TAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKRGRANLPIYPEDLSSE 194
EE+ II +H +GN+W+ +A LPGRTDNEIKN+WNT K+ + ED +
Sbjct: 316 APEEDDLIIRMHSLLGNRWSLIAGRLPGRTDNEIKNYWNTHLSKKLKIQ---GTEDTDTH 486
Query: 195 CLLNQENADMLTNEASQHDEAENKV--PEVIFKDYKLRPDILPPCFDKLLAGLLLRPSKR 252
+L + ++ + + + + K + + K + + PP L + +
Sbjct: 487 KMLENPQEEAASDGGNNNKKKKKKKNGGKKNKQKNKGKENDEPPKTQVYLPKPIRVKAMY 666
Query: 253 PWKSDMNLYSYSNNAAAPAAFDQLRKYPMPSPSSPPWEDIN---EPHSYNQFNEYDNPMV 309
++D N +++ +N+A+ + + + P+ S+ E N E + F+E D+ +V
Sbjct: 667 LQRTDSNTFTFDSNSASGSTSQEKEESPVTKESNVVSEVGNVGEESDGFGFFSE-DHDLV 843
Query: 310 GI 311
+
Sbjct: 844 NV 849
>TC211951 similar to GB|AAS10022.1|41619094|AY519552 MYB transcription factor
{Arabidopsis thaliana;} , partial (38%)
Length = 776
Score = 142 bits (359), Expect = 2e-34
Identities = 64/106 (60%), Positives = 82/106 (76%)
Frame = +2
Query: 76 LKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAIT 135
L+KG W+ EDE L+ ++ K+G G W+SV K GL RCGKSCRLRW N+LRPDL++GA +
Sbjct: 125 LRKGLWSPEEDEKLLNYITKHGHGCWSSVPKLAGLQRCGKSCRLRWINYLRPDLKRGAFS 304
Query: 136 AEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKRGR 181
+EE IIELH +GN+W+Q+AA LPGRTDNEIKN WN+ KK+ R
Sbjct: 305 QQEENSIIELHAVLGNRWSQIAAQLPGRTDNEIKNLWNSCLKKKLR 442
>TC216478 similar to UP|P93474 (P93474) Myb26, partial (83%)
Length = 925
Score = 142 bits (357), Expect = 4e-34
Identities = 62/103 (60%), Positives = 80/103 (77%)
Frame = +1
Query: 76 LKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAIT 135
++KGPWT ED IL+ ++ +GEG WNS+ K GL R GKSCRLRW N+LRPD+R+G IT
Sbjct: 100 VRKGPWTMEEDLILINYIANHGEGVWNSLAKAAGLKRTGKSCRLRWLNYLRPDVRRGNIT 279
Query: 136 AEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKK 178
EE+ I+ELH K GN+W+++A LPGRTDNEIKN+W TR +K
Sbjct: 280 PEEQLLIMELHAKWGNRWSKIAKHLPGRTDNEIKNYWRTRIQK 408
>TC221691 GmMYB6
Length = 590
Score = 142 bits (357), Expect = 4e-34
Identities = 68/148 (45%), Positives = 96/148 (63%)
Frame = +3
Query: 73 EGGLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKG 132
+ G +KG WT ED L+A+V +YG NW + + GLARCGKSCRLRW N+LRP++++G
Sbjct: 102 KSGTRKGTWTPEEDRKLIAYVTRYGSWNWRQLPRFAGLARCGKSCRLRWMNYLRPNVKRG 281
Query: 133 AITAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKRGRANLPIYPEDLS 192
T +E+ II +H K+GNKW+ +AA LPGRTDNEIKN W+T KK
Sbjct: 282 NFTQQEDECIIRMHKKLGNKWSAIAAELPGRTDNEIKNHWHTTLKK-------------- 419
Query: 193 SECLLNQENADMLTNEASQHDEAENKVP 220
+Q+NA +TNE ++ ++++KVP
Sbjct: 420 ----WSQQNA--ITNEEARTSKSKDKVP 485
>CA783329 homologue to GP|6651292|gb| myb-related transcription factor
{Pimpinella brachycarpa}, partial (48%)
Length = 443
Score = 142 bits (357), Expect = 4e-34
Identities = 61/105 (58%), Positives = 81/105 (77%)
Frame = +3
Query: 75 GLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAI 134
G+KKGPWTA ED+ L+ + G+ W +V K GL RCGKSCRLRW N+LRPDL++G +
Sbjct: 33 GVKKGPWTAEEDKKLINFILTNGQCCWRAVPKLAGLKRCGKSCRLRWTNYLRPDLKRGLL 212
Query: 135 TAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
T EE+ +I+LH ++GN+W+++AA LPGRTDNEIKN WNT KK+
Sbjct: 213 TEAEEQLVIDLHARLGNRWSKIAARLPGRTDNEIKNHWNTHIKKK 347
>TC235025 similar to UP|Q6R077 (Q6R077) MYB transcription factor, partial
(35%)
Length = 434
Score = 141 bits (355), Expect = 6e-34
Identities = 59/100 (59%), Positives = 78/100 (78%)
Frame = +1
Query: 75 GLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAI 134
G+KKGPWT EDE L+ ++ K+G G+W ++ K GL RCGKSCRLRW N+L PD+++G
Sbjct: 130 GVKKGPWTPEEDEKLIDYISKHGHGSWRTLPKRAGLNRCGKSCRLRWTNYLTPDIKRGKF 309
Query: 135 TAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNT 174
+ E+ER II LH +GNKW+++A LPGRTDNEIKN+WNT
Sbjct: 310 SEEDERIIINLHSVLGNKWSKIATHLPGRTDNEIKNYWNT 429
>TC229093 homologue to PIR|T51621|T51621 myb-like protein [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(44%)
Length = 964
Score = 141 bits (355), Expect = 6e-34
Identities = 61/105 (58%), Positives = 81/105 (77%)
Frame = +3
Query: 75 GLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAI 134
G+KKGPWT ED ILV+++Q++G GNW +V TGL+RC KSCRLRW N+LRP +++G
Sbjct: 180 GVKKGPWTPEEDIILVSYIQEHGPGNWKAVPANTGLSRCSKSCRLRWTNYLRPGIKRGNF 359
Query: 135 TAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
T +EE+ II L +GN+WA +AA LP RTDN+IKN+WNT KK+
Sbjct: 360 TDQEEKMIIHLQALLGNRWAAIAAYLPQRTDNDIKNYWNTYLKKK 494
>TC222952 homologue to UP|Q40173 (Q40173) Myb-related transcription factor,
partial (47%)
Length = 612
Score = 141 bits (355), Expect = 6e-34
Identities = 60/105 (57%), Positives = 82/105 (77%)
Frame = +2
Query: 75 GLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAI 134
GLKKGPWTA ED+ L++ + G+ W +V K GL RCGKSCRLRW N+LRPDL++G +
Sbjct: 155 GLKKGPWTAEEDKKLISFILTNGQCCWRAVPKLAGLLRCGKSCRLRWTNYLRPDLKRGLL 334
Query: 135 TAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
+ EE+ +I+LH ++GN+W+++A+ LPGRTDNEIKN WNT KK+
Sbjct: 335 SEYEEKMVIDLHAQLGNRWSKIASHLPGRTDNEIKNHWNTHIKKK 469
>BE352650
Length = 528
Score = 140 bits (354), Expect = 8e-34
Identities = 58/104 (55%), Positives = 80/104 (76%)
Frame = +1
Query: 76 LKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAIT 135
+KKGPWT EDE L+ ++ K+G G+W ++ K GL RCGKSCRLRW N+LRPD+++G +
Sbjct: 139 VKKGPWTPEEDEKLIDYISKHGHGSWRTLPKRAGLNRCGKSCRLRWTNYLRPDIKRGKFS 318
Query: 136 AEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKKR 179
++ER II H +GNKW+++AA LPGRTDNEIKN+W T +K+
Sbjct: 319 EDDERIIINFHSVLGNKWSKIAAHLPGRTDNEIKNYWTTHIRKK 450
>BF425254 homologue to PIR|S26605|S26 myb-related protein 1 - garden petunia,
partial (29%)
Length = 375
Score = 139 bits (350), Expect = 2e-33
Identities = 57/104 (54%), Positives = 80/104 (76%)
Frame = +3
Query: 75 GLKKGPWTAAEDEILVAHVQKYGEGNWNSVRKCTGLARCGKSCRLRWANHLRPDLRKGAI 134
GLKKGPWT ED+ L+A+++++G G+W ++ GL RCGKSCRLRW N+LRPD+++G
Sbjct: 27 GLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWTNYLRPDIKRGKF 206
Query: 135 TAEEERRIIELHHKMGNKWAQMAALLPGRTDNEIKNFWNTRCKK 178
+EE+ II+LH +GN+W+ ++ LP RTDNEIKN+WNT KK
Sbjct: 207 XMQEEQTIIQLHALLGNRWSSISTHLPKRTDNEIKNYWNTHLKK 338
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.312 0.131 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,139,608
Number of Sequences: 63676
Number of extensions: 335548
Number of successful extensions: 1971
Number of sequences better than 10.0: 200
Number of HSP's better than 10.0 without gapping: 1911
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1939
length of query: 416
length of database: 12,639,632
effective HSP length: 100
effective length of query: 316
effective length of database: 6,272,032
effective search space: 1981962112
effective search space used: 1981962112
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC148404.8


