

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
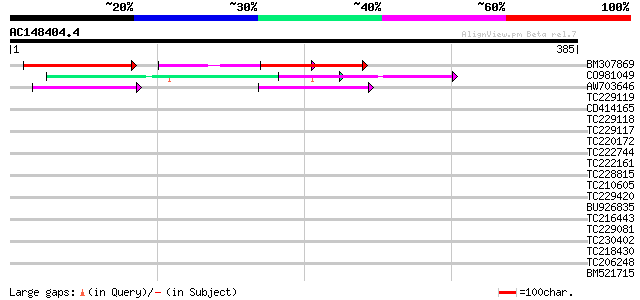
Query= AC148404.4 - phase: 0
(385 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BM307869 weakly similar to GP|20145851|emb auxin response factor... 135 3e-32
CO981049 74 8e-14
AW703646 64 9e-11
TC229119 38 0.007
CD414165 35 0.044
TC229118 similar to UP|Q6R2U8 (Q6R2U8) Reduced vernalization res... 35 0.057
TC229117 similar to UP|Q8L3W1 (Q8L3W1) Reduced vernalization res... 33 0.17
TC220172 similar to UP|Q8L3W1 (Q8L3W1) Reduced vernalization res... 33 0.22
TC222744 similar to UP|O80411 (O80411) Pharbitis kntted-like gen... 30 1.8
TC222161 weakly similar to UP|Q9SGD7 (Q9SGD7) T23G18.9, partial ... 30 2.4
TC228815 29 3.1
TC210605 weakly similar to UP|Q9FV01 (Q9FV01) SKP1 interacting p... 29 3.1
TC229420 homologue to GB|AAG30415.1|11118687|AF291666 homeodomai... 29 3.1
BU926835 29 4.1
TC216443 homologue to UP|Q08150 (Q08150) GTP-binding protein, co... 29 4.1
TC229081 UP|Q8LJR5 (Q8LJR5) Small GTP-binding protein, complete 29 4.1
TC230402 homologue to UP|Q08150 (Q08150) GTP-binding protein, co... 29 4.1
TC218430 28 5.3
TC206248 28 5.3
BM521715 28 5.3
>BM307869 weakly similar to GP|20145851|emb auxin response factor 36
{Arabidopsis thaliana}, partial (8%)
Length = 437
Score = 135 bits (340), Expect = 3e-32
Identities = 61/77 (79%), Positives = 68/77 (88%)
Frame = +2
Query: 10 INLLQALPKTFSDNLKKKLPGNVTLKGPSGVVWNVGLTARDDTIYFTNGWQRFVKDHSLK 69
IN L ALPK FSDNLKKKLP NVTLKGP GVVWN+G+T RDDT+YF +GW++FVKDH LK
Sbjct: 206 INTLLALPKAFSDNLKKKLPENVTLKGPGGVVWNIGMTTRDDTLYFAHGWEQFVKDHCLK 385
Query: 70 ENDFLVFKYNGESLFEV 86
ENDFLVFKYNGES F+V
Sbjct: 386 ENDFLVFKYNGESQFDV 436
Score = 128 bits (322), Expect = 4e-30
Identities = 57/73 (78%), Positives = 63/73 (86%)
Frame = +2
Query: 171 LALPKTFSDNLNKKLPENVTLKGPGGVAWKIGLITRDDTVYFLNGWQRFVNDHSLKDNDF 230
LALPK FSDNL KKLPENVTLKGPGGV W IG+ TRDDT+YF +GW++FV DH LK+NDF
Sbjct: 218 LALPKAFSDNLKKKLPENVTLKGPGGVVWNIGMTTRDDTLYFAHGWEQFVKDHCLKENDF 397
Query: 231 LVFNYNGESHFDV 243
LVF YNGES FDV
Sbjct: 398 LVFKYNGESQFDV 436
Score = 52.4 bits (124), Expect = 3e-07
Identities = 36/107 (33%), Positives = 50/107 (46%)
Frame = +1
Query: 102 FVGKCGQAHTEQVCNKGKSSNNSVEEVSTPSNGSVECSLPEKSWEEDIYWTHFQFIHFTQ 161
F G+ + T + CN S E V G +SWEEDIYW+HFQF+HF Q
Sbjct: 28 FPGRPTRPQTTKFCNFFSSLIVVGEMVGQNCGGC-------RSWEEDIYWSHFQFLHFVQ 186
Query: 162 LLPEDFKQQLALPKTFSDNLNKKLPENVTLKGPGGVAWKIGLITRDD 208
L D+ Q L + S+++ + E V K +W + RDD
Sbjct: 187 FLRADYDQHLV---STSESIFR*FKEEVAGKCDP*GSWWCCVEYRDD 318
>CO981049
Length = 665
Score = 74.3 bits (181), Expect = 8e-14
Identities = 40/122 (32%), Positives = 57/122 (45%)
Frame = -2
Query: 183 KKLPENVTLKGPGGVAWKIGLITRDDTVYFLNGWQRFVNDHSLKDNDFLVFNYNGESHFD 242
K L + LK P G W++ L RD V+F GW++FV HSL LVF Y+G HF
Sbjct: 652 KHLSNTMFLKLPNGAEWRVNLEKRDGRVWFQEGWKKFVEHHSLAHGHLLVFKYDGTFHFH 473
Query: 243 VLIFDGESFCEKEASYFVGKCSHTQTELGGSKANETNNSIEEVNTASSDGGVECGLYEKF 302
VLIFD + E Y V K +H + + + N S+ + ++K
Sbjct: 472 VLIFDPSA---NEIDYPVNKANHKRVRISSEEIQPPTTCKTSGNKRSNSNLQDNAFHQKV 302
Query: 303 QD 304
+D
Sbjct: 301 RD 296
Score = 67.4 bits (163), Expect = 1e-11
Identities = 59/215 (27%), Positives = 85/215 (39%), Gaps = 13/215 (6%)
Frame = -2
Query: 26 KKLPGNVTLKGPSGVVWNVGLTARDDTIYFTNGWQRFVKDHSLKENDFLVFKYNGESLFE 85
K L + LK P+G W V L RD ++F GW++FV+ HSL LVFKY+G F
Sbjct: 652 KHLSNTMFLKLPNGAEWRVNLEKRDGRVWFQEGWKKFVEHHSLAHGHLLVFKYDGTFHFH 473
Query: 86 VLIFHGDSFCEKAASYFVGKCG-----------QAHTEQVCNKGKSSNNSVEEVSTPSNG 134
VLIF + Y V K Q T + K SN+++++ +
Sbjct: 472 VLIFDPSA---NEIDYPVNKANHKRVRISSEEIQPPTTCKTSGNKRSNSNLQDNAFHQKV 302
Query: 135 SVECSLPEKSWEEDIYWTHFQFIHFTQLLPEDFKQQLALPKTFSDNLNKKLPENVTLKGP 194
E E I FT + KQ + LPK K + V L
Sbjct: 301 RDHKGRYESPSEGKRNMEAAGSISFTVRMKSSSKQHMYLPKDSLKGYIKGGEQYVKLL-V 125
Query: 195 GGVAWKIGLI--TRDDTVYFLNGWQRFVNDHSLKD 227
G +W++ L+ + +F W F LK+
Sbjct: 124 GERSWRVKLVHYKNRSSCFFSANWPAFARXXDLKE 20
>AW703646
Length = 422
Score = 64.3 bits (155), Expect = 9e-11
Identities = 31/78 (39%), Positives = 41/78 (51%)
Frame = +2
Query: 170 QLALPKTFSDNLNKKLPENVTLKGPGGVAWKIGLITRDDTVYFLNGWQRFVNDHSLKDND 229
+L +P F K+L + LK P G WK+ L RD ++F GW+ F HSL
Sbjct: 32 KLMIPNKFVKKYGKRLQNTLFLKTPNGAEWKMILKKRDGKIWFQKGWKEFAEYHSLAHGH 211
Query: 230 FLVFNYNGESHFDVLIFD 247
LVF ++ SHF V IFD
Sbjct: 212 LLVFRWDVTSHFQVHIFD 265
Score = 57.4 bits (137), Expect = 1e-08
Identities = 28/74 (37%), Positives = 40/74 (53%)
Frame = +2
Query: 16 LPKTFSDNLKKKLPGNVTLKGPSGVVWNVGLTARDDTIYFTNGWQRFVKDHSLKENDFLV 75
+P F K+L + LK P+G W + L RD I+F GW+ F + HSL LV
Sbjct: 41 IPNKFVKKYGKRLQNTLFLKTPNGAEWKMILKKRDGKIWFQKGWKEFAEYHSLAHGHLLV 220
Query: 76 FKYNGESLFEVLIF 89
F+++ S F+V IF
Sbjct: 221 FRWDVTSHFQVHIF 262
>TC229119
Length = 455
Score = 38.1 bits (87), Expect = 0.007
Identities = 18/36 (50%), Positives = 23/36 (63%)
Frame = +2
Query: 55 FTNGWQRFVKDHSLKENDFLVFKYNGESLFEVLIFH 90
F +GWQ FV+ +S+ FLVF Y G S F V IF+
Sbjct: 5 FVDGWQDFVQHYSIGVGYFLVFMYEGNSSFIVHIFN 112
Score = 37.7 bits (86), Expect = 0.009
Identities = 18/36 (50%), Positives = 23/36 (63%)
Frame = +2
Query: 212 FLNGWQRFVNDHSLKDNDFLVFNYNGESHFDVLIFD 247
F++GWQ FV +S+ FLVF Y G S F V IF+
Sbjct: 5 FVDGWQDFVQHYSIGVGYFLVFMYEGNSSFIVHIFN 112
>CD414165
Length = 623
Score = 35.4 bits (80), Expect = 0.044
Identities = 28/116 (24%), Positives = 44/116 (37%), Gaps = 4/116 (3%)
Frame = -1
Query: 135 SVECSLPEKSWEEDIYWTHFQFIHFTQLLPEDFKQQLALPKTFSDNLNKKLPENVTLKGP 194
S C L E + +W H T+ ED L + F+ ++ + + L
Sbjct: 482 SEACKLAESFTSRNPHWKHLM----TKCNVEDHCT-LHIATQFACKYIPEVVKQIILWNS 318
Query: 195 GGVAWKIGLIT----RDDTVYFLNGWQRFVNDHSLKDNDFLVFNYNGESHFDVLIF 246
G W++ + F GW +FV D+ L D +F E+H V IF
Sbjct: 317 EGKFWEVEVTCLRYPNKRYTRFTTGWGKFVRDNKLMPGDTCIFELEDENHMSVHIF 150
>TC229118 similar to UP|Q6R2U8 (Q6R2U8) Reduced vernalization response 1,
partial (17%)
Length = 470
Score = 35.0 bits (79), Expect = 0.057
Identities = 18/62 (29%), Positives = 30/62 (48%)
Frame = +3
Query: 16 LPKTFSDNLKKKLPGNVTLKGPSGVVWNVGLTARDDTIYFTNGWQRFVKDHSLKENDFLV 75
LP TF++ + G + L+ +G W+V R + GW F +++L E D V
Sbjct: 15 LPSTFAEKNLNGVSGFIKLQLSNGRQWSVRCLYRGGRAKLSQGWFEFTVENNLGEGDVCV 194
Query: 76 FK 77
F+
Sbjct: 195FE 200
Score = 29.3 bits (64), Expect = 3.1
Identities = 15/61 (24%), Positives = 26/61 (42%)
Frame = +3
Query: 173 LPKTFSDNLNKKLPENVTLKGPGGVAWKIGLITRDDTVYFLNGWQRFVNDHSLKDNDFLV 232
LP TF++ + + L+ G W + + R GW F +++L + D V
Sbjct: 15 LPSTFAEKNLNGVSGFIKLQLSNGRQWSVRCLYRGGRAKLSQGWFEFTVENNLGEGDVCV 194
Query: 233 F 233
F
Sbjct: 195 F 197
>TC229117 similar to UP|Q8L3W1 (Q8L3W1) Reduced vernalization response 1,
partial (38%)
Length = 904
Score = 33.5 bits (75), Expect = 0.17
Identities = 20/76 (26%), Positives = 35/76 (45%), Gaps = 2/76 (2%)
Frame = +3
Query: 16 LPKTFSDNLKKKLPGNVTLKGPSGVVWNVGLTARDDTIYFTNGWQRFVKDHSLKENDFLV 75
LP F++ + G + L+ +G W+V R + GW F +++L E D V
Sbjct: 426 LPSCFAEKNLNGVSGFIKLQLSNGRQWSVRCLYRGGRAKLSQGWFEFTVENNLGEGDVCV 605
Query: 76 FKY--NGESLFEVLIF 89
F+ E + +V +F
Sbjct: 606 FELLRTKEVVLQVTVF 653
>TC220172 similar to UP|Q8L3W1 (Q8L3W1) Reduced vernalization response 1,
partial (39%)
Length = 798
Score = 33.1 bits (74), Expect = 0.22
Identities = 21/77 (27%), Positives = 35/77 (45%), Gaps = 2/77 (2%)
Frame = +3
Query: 16 LPKTFSDNLKKKLPGNVTLKGPSGVVWNVGLTARDDTIYFTNGWQRFVKDHSLKENDFLV 75
LP F++ + G + L+ +G W V + + GW F +++L E D V
Sbjct: 204 LPSCFAEKHLNGVSGFIKLQISNGRQWPVRCLYKGGRAKLSQGWFEFSLENNLGEGDVCV 383
Query: 76 FKY--NGESLFEVLIFH 90
F+ E + +V IFH
Sbjct: 384 FELLRMKEVVLQVTIFH 434
>TC222744 similar to UP|O80411 (O80411) Pharbitis kntted-like gene 3, partial
(38%)
Length = 781
Score = 30.0 bits (66), Expect = 1.8
Identities = 16/53 (30%), Positives = 29/53 (54%), Gaps = 3/53 (5%)
Frame = +1
Query: 255 EASYFVGKCSHTQTELGG---SKANETNNSIEEVNTASSDGGVECGLYEKFQD 304
EA+ F+G + L +K+++ NN +EV + +S+ + CG E F+D
Sbjct: 25 EATLFLGDMESQLSNLCNETLTKSSDNNNRSDEVASGASEEELSCGEMEAFED 183
>TC222161 weakly similar to UP|Q9SGD7 (Q9SGD7) T23G18.9, partial (52%)
Length = 978
Score = 29.6 bits (65), Expect = 2.4
Identities = 11/38 (28%), Positives = 23/38 (59%), Gaps = 1/38 (2%)
Frame = -1
Query: 115 CNKGKSS-NNSVEEVSTPSNGSVECSLPEKSWEEDIYW 151
C G+S+ NN+++ + S G ++C + E W+ ++ W
Sbjct: 765 CQCGESNDNNNIQPENGASLGWLKCHIEENHWQNEVKW 652
>TC228815
Length = 765
Score = 29.3 bits (64), Expect = 3.1
Identities = 13/31 (41%), Positives = 18/31 (57%)
Frame = -3
Query: 341 VTKTTAIQFPYQPTGKRSKRRRSSTSANCCW 371
+ KT AI+F P +R+ RRR+ T CW
Sbjct: 286 LVKTRAIRFGSDPGERRNSRRRAPTRICGCW 194
>TC210605 weakly similar to UP|Q9FV01 (Q9FV01) SKP1 interacting partner 4,
partial (12%)
Length = 661
Score = 29.3 bits (64), Expect = 3.1
Identities = 14/41 (34%), Positives = 23/41 (55%)
Frame = +1
Query: 102 FVGKCGQAHTEQVCNKGKSSNNSVEEVSTPSNGSVECSLPE 142
F+ + T + +KGK S+NS+ EV +N + C LP+
Sbjct: 100 FLTRSSVPFTMEHADKGKESSNSINEVEV-TNSPIICGLPD 219
>TC229420 homologue to GB|AAG30415.1|11118687|AF291666 homeodomain-containing
transcription factor Nkx6.1 {Mus musculus;} , partial
(8%)
Length = 870
Score = 29.3 bits (64), Expect = 3.1
Identities = 12/26 (46%), Positives = 17/26 (65%)
Frame = +2
Query: 56 TNGWQRFVKDHSLKENDFLVFKYNGE 81
T GW+ F D++L+E D VFK G+
Sbjct: 419 TGGWKHFSLDNNLEEGDACVFKPAGQ 496
>BU926835
Length = 451
Score = 28.9 bits (63), Expect = 4.1
Identities = 11/24 (45%), Positives = 14/24 (57%)
Frame = -2
Query: 214 NGWQRFVNDHSLKDNDFLVFNYNG 237
NGWQ +ND S+K +NY G
Sbjct: 219 NGWQSHLNDSSVKGYQLAFWNYQG 148
>TC216443 homologue to UP|Q08150 (Q08150) GTP-binding protein, complete
Length = 1277
Score = 28.9 bits (63), Expect = 4.1
Identities = 23/88 (26%), Positives = 40/88 (45%), Gaps = 4/88 (4%)
Frame = +3
Query: 204 ITRDDTVYFLNGWQRFVNDHSLKDNDFLVFNYNGESHFDVLIF----DGESFCEKEASYF 259
+TR T ++ W + + +H+ D + +V +S L+ DG+S+ EKE+ YF
Sbjct: 642 VTRHATFENVDRWLKELRNHT--DANIVVMLVGNKSDLRHLVAVSTEDGKSYAEKESLYF 815
Query: 260 VGKCSHTQTELGGSKANETNNSIEEVNT 287
+ E +A N+ EV T
Sbjct: 816 M--------ETSALEATNVENAFAEVLT 875
>TC229081 UP|Q8LJR5 (Q8LJR5) Small GTP-binding protein, complete
Length = 1478
Score = 28.9 bits (63), Expect = 4.1
Identities = 24/86 (27%), Positives = 37/86 (42%), Gaps = 4/86 (4%)
Frame = +1
Query: 204 ITRDDTVYFLNGWQRFVNDHSLKDNDFLVFNYNGESHFDVLIF----DGESFCEKEASYF 259
+TR T W + + DH+ D + +V +S L+ DG+SF EKE+ YF
Sbjct: 655 VTRRATFENAARWLKELRDHT--DPNIVVMLIGNKSDLRHLVAVPTEDGKSFAEKESLYF 828
Query: 260 VGKCSHTQTELGGSKANETNNSIEEV 285
+ E +A N+ EV
Sbjct: 829 M--------ETSALEATNVENAFTEV 882
>TC230402 homologue to UP|Q08150 (Q08150) GTP-binding protein, complete
Length = 844
Score = 28.9 bits (63), Expect = 4.1
Identities = 23/88 (26%), Positives = 40/88 (45%), Gaps = 4/88 (4%)
Frame = +1
Query: 204 ITRDDTVYFLNGWQRFVNDHSLKDNDFLVFNYNGESHFDVLIF----DGESFCEKEASYF 259
+TR T ++ W + + +H+ D + +V +S L+ DG+S+ EKE+ YF
Sbjct: 370 VTRHATFENVDRWLKELRNHT--DANIVVMLVGNKSDLRHLVAVSTEDGKSYAEKESLYF 543
Query: 260 VGKCSHTQTELGGSKANETNNSIEEVNT 287
+ E +A N+ EV T
Sbjct: 544 M--------ETSALEATNVENAFAEVLT 603
>TC218430
Length = 1397
Score = 28.5 bits (62), Expect = 5.3
Identities = 13/45 (28%), Positives = 27/45 (59%)
Frame = +1
Query: 112 EQVCNKGKSSNNSVEEVSTPSNGSVECSLPEKSWEEDIYWTHFQF 156
++V + K++ S E+ S + G++ +K+W+ D+YW H+ F
Sbjct: 136 DEVSYECKAAYPSFEDSSRIALGNI-LKFRKKNWQFDLYWWHYIF 267
>TC206248
Length = 1008
Score = 28.5 bits (62), Expect = 5.3
Identities = 20/83 (24%), Positives = 35/83 (42%)
Frame = +1
Query: 120 SSNNSVEEVSTPSNGSVECSLPEKSWEEDIYWTHFQFIHFTQLLPEDFKQQLALPKTFSD 179
SS NS+++V +NG S + EE ++H P+ +P+
Sbjct: 577 SSTNSIQQVIESNNGLHGNSSKAMNMEEKFCYSH----------PQKLLVDAEIPRFGLS 726
Query: 180 NLNKKLPENVTLKGPGGVAWKIG 202
N + N+T +GP ++ IG
Sbjct: 727 NQPRDTGSNITCEGPIKTSFMIG 795
>BM521715
Length = 277
Score = 28.5 bits (62), Expect = 5.3
Identities = 16/60 (26%), Positives = 24/60 (39%), Gaps = 1/60 (1%)
Frame = +2
Query: 157 IHFTQL-LPEDFKQQLALPKTFSDNLNKKLPENVTLKGPGGVAWKIGLITRDDTVYFLNG 215
+HF ++ L + LPK F+ + V LK G WKI ++F G
Sbjct: 98 VHFVKIILTTSLADGILLPKKFTRKYGDGMSNPVFLKPADGTEWKIHYTKHGGEIWFQKG 277
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.133 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,948,103
Number of Sequences: 63676
Number of extensions: 274986
Number of successful extensions: 1425
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 1363
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1425
length of query: 385
length of database: 12,639,632
effective HSP length: 99
effective length of query: 286
effective length of database: 6,335,708
effective search space: 1812012488
effective search space used: 1812012488
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC148404.4


