

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148403.7 - phase: 0
(315 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
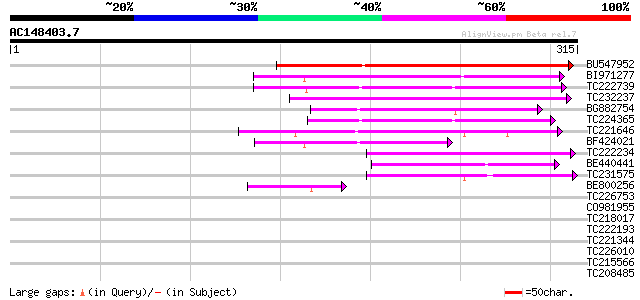
Score E
Sequences producing significant alignments: (bits) Value
BU547952 127 5e-30
BI971277 118 3e-27
TC222739 89 3e-18
TC232237 89 3e-18
BG882754 80 9e-16
TC224365 73 2e-13
TC221646 72 3e-13
BF424021 70 1e-12
TC222234 69 3e-12
BE440441 55 3e-08
TC231575 55 5e-08
BE800256 48 5e-06
TC226753 similar to UP|Q9XIN0 (Q9XIN0) Expressed protein (BHLH t... 35 0.044
CO981955 32 0.49
TC218017 similar to GB|AAF82809.1|9082326|AF283277 polyadenylati... 30 1.1
TC222193 UP|Q39873 (Q39873) Lea protein precursor, complete 30 1.1
TC221344 30 1.4
TC226010 weakly similar to UP|Q8YPJ4 (Q8YPJ4) 50S ribosomal prot... 30 1.9
TC215566 similar to UP|Q9FSG4 (Q9FSG4) Wound-induced GSK-3-like ... 30 1.9
TC208485 similar to UP|Q96424 (Q96424) O-methyltransferase, part... 28 7.1
>BU547952
Length = 699
Score = 127 bits (320), Expect = 5e-30
Identities = 70/166 (42%), Positives = 100/166 (60%), Gaps = 1/166 (0%)
Frame = -3
Query: 149 KKSKTRSLKDVSFNRGVLRLPKLVVDDHTECMFLNLIAFELLHVDGTRKEVISFVCFIDT 208
KK KT D+ F G LR+P+L++ D T+ +FLNLIA E H+D + ++ S+V F+D
Sbjct: 697 KKRKTDRXWDIKFKDGKLRIPRLLIHDGTKSLFLNLIAXEQCHLDCSN-DITSYVIFMDN 521
Query: 209 IIDSAVDVAILIRSGIIINYLESDKAVAKLFNSLAKEIPMD-REGELEEVTKSMISYCKK 267
+I+S DV L GII ++L SD VA LFN L +E+ D L +++ + Y
Sbjct: 520 LINSPEDVGYLHYRGIIEHWLGSDAEVADLFNRLCQEVVFDINNSYLSPLSEDVNRYYNH 341
Query: 268 PWKSWRASLIQTYFRNPWAMVSLVAAFFLFALTIIQTIYTVGQFYQ 313
W +W ASL YF NPWA++SLVAA L LT+ QT Y + +Y+
Sbjct: 340 RWNTWCASLRHNYFSNPWAIISLVAAVVLLLLTLAQTYYGIYGYYR 203
>BI971277
Length = 700
Score = 118 bits (296), Expect = 3e-27
Identities = 71/177 (40%), Positives = 103/177 (58%), Gaps = 4/177 (2%)
Frame = -1
Query: 136 RSAMELKEAGISFKKSKTRSLKDVSFN----RGVLRLPKLVVDDHTECMFLNLIAFELLH 191
R+ ELKEAGI K SKTR +DVSF+ R L+LP++VVDD T LNLIA+E+
Sbjct: 685 RNIKELKEAGIVLKSSKTRRPRDVSFSYGWIRSELKLPEIVVDDTTAATVLNLIAYEMCP 506
Query: 192 VDGTRKEVISFVCFIDTIIDSAVDVAILIRSGIIINYLESDKAVAKLFNSLAKEIPMDRE 251
+ S+V F+D++ID DV L I++N L SD+ VA LFN+++ ++ D
Sbjct: 505 DFENDYGICSYVSFLDSLIDHPDDVKALRSEQILLNSLGSDEEVANLFNTISTDLVPDMV 326
Query: 252 GELEEVTKSMISYCKKPWKSWRASLIQTYFRNPWAMVSLVAAFFLFALTIIQTIYTV 308
+ +V + + ++W A TYF NPWA+++ AA ALT +QT YT+
Sbjct: 325 -KYADVRNEIEKHYSDKSRTWLALGYHTYFSNPWAIIAFHAAVVGLALTFVQTWYTI 158
>TC222739
Length = 756
Score = 88.6 bits (218), Expect = 3e-18
Identities = 56/194 (28%), Positives = 92/194 (46%), Gaps = 20/194 (10%)
Frame = +2
Query: 136 RSAMELKEAGISFKKSKTRSLKDVSFNR--------------------GVLRLPKLVVDD 175
R+A +L EAG+SF+K + RS D+ F + L++P L VD
Sbjct: 158 RTATKLNEAGVSFEKVQGRSYLDIKFEKTPILSWFLCFGCLPFSKWFKARLQIPHLKVDQ 337
Query: 176 HTECMFLNLIAFELLHVDGTRKEVISFVCFIDTIIDSAVDVAILIRSGIIINYLESDKAV 235
TEC+ NLIA E H + + ++V ID++I + DV +L+ + II + L S +
Sbjct: 338 VTECVLRNLIALEQCHYSD-QPFICNYVTLIDSLIHTQEDVELLVDTEIIDHELGSHTEL 514
Query: 236 AKLFNSLAKEIPMDREGELEEVTKSMISYCKKPWKSWRASLIQTYFRNPWAMVSLVAAFF 295
A + N L K + + + TK + + WK + LI YFR+PW S +
Sbjct: 515 ATMINGLCKHV-LVTSNYYGKTTKELNEHYNCCWKHYMGMLISVYFRDPWRFSSTIVGIA 691
Query: 296 LFALTIIQTIYTVG 309
+F ++ + +G
Sbjct: 692 VFLFAVVNFLRIIG 733
>TC232237
Length = 673
Score = 88.6 bits (218), Expect = 3e-18
Identities = 50/159 (31%), Positives = 84/159 (52%), Gaps = 2/159 (1%)
Frame = +1
Query: 156 LKDVSFN-RGVLRLPKLVVDDHTECMFLNLIAFELLHVDGTRKEVISFVCFIDTIIDSAV 214
L D++++ GVL +P L + D +E F N++AFE H+ + + +D +I++
Sbjct: 19 LLDMTYSDEGVLTMPILNIADDSEMFFRNIVAFEQCHLSDDTNIITQYRKILDFLINTEK 198
Query: 215 DVAILIRSGIIINYLESDKAVAKLFNSLAKEIPMDREGELE-EVTKSMISYCKKPWKSWR 273
DV +L+ II+N++ AVA + NSL I M R + + S+ + + P ++
Sbjct: 199 DVNVLVDKKIIVNWMGDANAVATMVNSLGSNIGMPRFNPVYFSLCNSLNDFYESPCNKYK 378
Query: 274 ASLIQTYFRNPWAMVSLVAAFFLFALTIIQTIYTVGQFY 312
A YF PW + S VAA L LT+IQTI ++ +
Sbjct: 379 AIFKHDYFNTPWKIASTVAAIVLLLLTLIQTICSINSLF 495
>BG882754
Length = 399
Score = 80.5 bits (197), Expect = 9e-16
Identities = 42/131 (32%), Positives = 76/131 (57%), Gaps = 2/131 (1%)
Frame = +2
Query: 168 LPKLVVDDHTECMFLNLIAFELLHVDGTRKEVISFVCFIDTIIDSAVDVAILIRSGIIIN 227
+P+LVVDD TE +F N++A E H + + +V F+D ++++ DV IL++ + IN
Sbjct: 5 IPQLVVDDLTETLFRNMVALEQCHYL-FQSYITDYVGFLDFLVNTNRDVDILVQERVFIN 181
Query: 228 YLESDKAVAKLFNSLAKEI--PMDREGELEEVTKSMISYCKKPWKSWRASLIQTYFRNPW 285
+L +VA + N L K I P++ + +V++ + ++ K PW+ +++L + Y R PW
Sbjct: 182 WLGDTDSVATMINGLMKNITTPINTSSQYLDVSEKLNAFHKNPWRKLKSALRRDYCRGPW 361
Query: 286 AMVSLVAAFFL 296
+ AA L
Sbjct: 362 QTAASTAAVIL 394
>TC224365
Length = 766
Score = 72.8 bits (177), Expect = 2e-13
Identities = 43/138 (31%), Positives = 68/138 (49%)
Frame = +1
Query: 166 LRLPKLVVDDHTECMFLNLIAFELLHVDGTRKEVISFVCFIDTIIDSAVDVAILIRSGII 225
LR+P+L VD TE +F NL+AFE H + ++V FID++I + +DV +L+ +I
Sbjct: 109 LRIPQLKVDHTTERVFKNLVAFEQFHYPD-KPYFCNYVSFIDSLIHTQLDVELLVEKEVI 285
Query: 226 INYLESDKAVAKLFNSLAKEIPMDREGELEEVTKSMISYCKKPWKSWRASLIQTYFRNPW 285
++ L SDK VA L N L K + + + + + W A+L YFR+ W
Sbjct: 286 VHELGSDKEVATLVNGLCKHV-VTNSTCYHHIINKLNDHYMNDWNHTIAALRLVYFRDLW 462
Query: 286 AMVSLVAAFFLFALTIIQ 303
V + + Q
Sbjct: 463 RASGTVVGIAVLVFAVFQ 516
>TC221646
Length = 790
Score = 72.0 bits (175), Expect = 3e-13
Identities = 56/209 (26%), Positives = 99/209 (46%), Gaps = 29/209 (13%)
Frame = +1
Query: 128 KEDEDHLNRSAMELKEAGISFKKSKTRSLK------DVSFNRGVLRLPKLVVDDHTECMF 181
K+ + L R A L+ AG++ +K + S D+ F++GVL +P L V D TE
Sbjct: 64 KQAQQELKRCATRLRAAGVTIRKVERHSKLVNWFGFDIRFSKGVLEIPPLHVVDTTEVYL 243
Query: 182 LNLIAFELLHVDGTRKEVISFVCFIDTIIDSAVDVAILIRSGIIINYLE-SDKAVAKLFN 240
N IA+E + G ++ S+ F+ ++ S D+ +L+ +G+++ + S++ + LF
Sbjct: 244 RNFIAWEQSRI-GINRQFTSYALFLRGLMCSVQDIELLVENGVLVKGTKISNRDLLTLFG 420
Query: 241 SLAKEIP-MDRE--------------GELEEVTKSMISYCKKPWKSWRAS-------LIQ 278
++ K + MD L + M+ C + R S LI+
Sbjct: 421 TITKGVDQMDNSYSKLCEDLNAYSAVNPLRKFPILMLHCCNLCVECIRYSCKHSYKILIR 600
Query: 279 TYFRNPWAMVSLVAAFFLFALTIIQTIYT 307
+ N W ++ +V A L ALTI+QT Y+
Sbjct: 601 DHIPNVWKLIGIVVAAALLALTIMQTYYS 687
>BF424021
Length = 421
Score = 70.5 bits (171), Expect = 1e-12
Identities = 38/112 (33%), Positives = 65/112 (57%), Gaps = 2/112 (1%)
Frame = +3
Query: 137 SAMELKEAGISFKKSKTRSLKDVSFN--RGVLRLPKLVVDDHTECMFLNLIAFELLHVDG 194
SA +L E G+ FK S + L ++ ++ +GV+ +P L ++D TE +F N++A E H
Sbjct: 75 SASQLSEVGMVFKASSCKCLFELKYHHRKGVMEMPCLTIEDRTETLFRNILALEQCHYI- 251
Query: 195 TRKEVISFVCFIDTIIDSAVDVAILIRSGIIINYLESDKAVAKLFNSLAKEI 246
V F+ ++ +I++ DV IL+ II+N++ AV K+FNSL +
Sbjct: 252 LSPNVTQFLFLLNFLINTEKDVNILVDKKIIVNWMGDANAVVKMFNSLCSNV 407
>TC222234
Length = 580
Score = 68.9 bits (167), Expect = 3e-12
Identities = 30/117 (25%), Positives = 68/117 (57%), Gaps = 1/117 (0%)
Frame = +3
Query: 199 VISFVCFIDTIIDSAVDVAILIRSGIIINYLESDKAVAKLFNSLAKEIP-MDREGELEEV 257
V +V +D +++++ DV +L+R G+++N+L +VA +FN L K + ++ E+
Sbjct: 24 VTDYVAVLDFLVNTSRDVDVLVRKGVLVNWLGDSDSVADMFNGLWKNVTHINFSSHYSEL 203
Query: 258 TKSMISYCKKPWKSWRASLIQTYFRNPWAMVSLVAAFFLFALTIIQTIYTVGQFYQK 314
+ + ++C+ P +++L + Y + PW + +A L L+++Q++ +V Q Q+
Sbjct: 204 CQKLNAFCRNPCHKLKSTLRRDYCKTPWQIAVSIAGIVLLVLSLVQSVCSVLQVIQQ 374
>BE440441
Length = 427
Score = 55.5 bits (132), Expect = 3e-08
Identities = 31/104 (29%), Positives = 52/104 (49%)
Frame = -1
Query: 202 FVCFIDTIIDSAVDVAILIRSGIIINYLESDKAVAKLFNSLAKEIPMDREGELEEVTKSM 261
+V ID++I + +DV +L+ +I++ L SDK VA L N L+K + + E + +
Sbjct: 427 YVSLIDSLIHTQLDVELLVEKEVIVHELGSDKEVAVLVNGLSKHVVANTTCYYETINELN 248
Query: 262 ISYCKKPWKSWRASLIQTYFRNPWAMVSLVAAFFLFALTIIQTI 305
Y + W A+L YFR+PW S + + + Q I
Sbjct: 247 KHY-QNIWNRTMAALWLVYFRDPWRASSTLVGIVVLIFAVFQFI 119
>TC231575
Length = 527
Score = 54.7 bits (130), Expect = 5e-08
Identities = 30/119 (25%), Positives = 55/119 (46%), Gaps = 2/119 (1%)
Frame = +2
Query: 199 VISFVCFIDTIIDSAVDVAILIRSGIIINYLESDKAVAKLFNSLAKEIPMDRE--GELEE 256
+ ++V ID++I + DV +L+ II++ L +A + N L K + ++ G+
Sbjct: 17 ICNYVSLIDSLIHTQEDVELLVDKEIIVHELGCHNELATMINGLCKHVVVNCNYYGKTSR 196
Query: 257 VTKSMISYCKKPWKSWRASLIQTYFRNPWAMVSLVAAFFLFALTIIQTIYTVGQFYQKD 315
+ C WK + L YFR+PW + S V +F I+ + ++ KD
Sbjct: 197 KLNDHYNCC---WKHYMGMLRSVYFRDPWRLSSTVVGVVIFLFAIVNFLRVTNLYHPKD 364
>BE800256
Length = 403
Score = 48.1 bits (113), Expect = 5e-06
Identities = 27/75 (36%), Positives = 42/75 (56%), Gaps = 20/75 (26%)
Frame = +2
Query: 133 HLNRSAMELKEAGISFKKSKTRSLKDVSFNRGVL--------------------RLPKLV 172
H+ R+A +L+++G+SF+K R L D+SF++ + R+P+L
Sbjct: 179 HVLRTATKLQDSGVSFEKDVERRLLDISFDKKPILSSFLCFGCLPYLNHFKARFRIPQLK 358
Query: 173 VDDHTECMFLNLIAF 187
VD TEC+F NLIAF
Sbjct: 359 VDHTTECVFRNLIAF 403
>TC226753 similar to UP|Q9XIN0 (Q9XIN0) Expressed protein (BHLH transcription
factor), partial (30%)
Length = 1772
Score = 35.0 bits (79), Expect = 0.044
Identities = 38/185 (20%), Positives = 85/185 (45%), Gaps = 16/185 (8%)
Frame = +3
Query: 1 MELKYTGINFETSETISLEVSEEDEDHLNRSAMELKYTGINFKSSETISLEVSEE----- 55
M+LK +N ++ ++ + S+++ +HL++ A + G+ +S +++ +SE
Sbjct: 36 MDLKRRLLNGSWNKLLATD-SDDNTEHLDKKATCMNLQGVGPDNSYSVNEAISESGIFSG 212
Query: 56 -DEDHL-------NQSAMKLKETRINFKSSETSLEVSEEDKDDIIELVIEDHEHALPQSA 107
D DHL QSA K ++ +++ T + + + + V+ DH P+
Sbjct: 213 TDTDHLLDAVVSKAQSAAKQNYDEMSCRTTLTRISTASIPSP-VCKQVMPDH--VAPRGL 383
Query: 108 MVVKKTGINFKTSETS---LKVSKEDEDHLNRSAMELKEAGISFKKSKTRSLKDVSFNRG 164
KTG+ ++ETS SK+D + +++ S+ ++ + ++ S + G
Sbjct: 384 FDFPKTGVKTASAETSSLRSGCSKDDAGNCSQTTSIYGSKLSSWVENSSNFKRESSVSTG 563
Query: 165 VLRLP 169
+ P
Sbjct: 564 YSKRP 578
>CO981955
Length = 669
Score = 31.6 bits (70), Expect = 0.49
Identities = 49/182 (26%), Positives = 73/182 (39%), Gaps = 33/182 (18%)
Frame = -1
Query: 160 SFNRGVLRLPKLVVDDHTECMFLNLIAFELLHVDGTRKE---------------VISFVC 204
S+N GVL +P L + TE + + IA+E H + ++ E V +F+
Sbjct: 588 SYNNGVLLIPHLRITQTTEPKWRSFIAWE-HHRNKSKNEEFGNSTEQDGIEYANVFTFLA 412
Query: 205 FI-DTIIDSAVDVAILIRSGIIINYL-ESDKAVAKLFNSLAKEIPMDREGE--------- 253
+ + +I A DV IL GII + L S+ V F S+A I G
Sbjct: 411 LLFNDLICCASDVQILKNKGIIKDELGMSNHEVVDFFGSVANGIDRGHVGSGYSNIIDAL 232
Query: 254 -----LEEVTKSMISYCKKPWK--SWRASLIQTYFRNPWAMVSLVAAFFLFALTIIQTIY 306
VT+ I C + W L Q + R + AA + LT++QT Y
Sbjct: 231 NTFSATNFVTRFPIIACHDLSRITEWIYGL-QKFLRRGYNF----AAALITLLTVVQTCY 67
Query: 307 TV 308
V
Sbjct: 66 AV 61
>TC218017 similar to GB|AAF82809.1|9082326|AF283277 polyadenylation
cleavage/specificity factor 100 kDa subunit {Arabidopsis
thaliana;} , partial (35%)
Length = 1292
Score = 30.4 bits (67), Expect = 1.1
Identities = 18/45 (40%), Positives = 26/45 (57%)
Frame = -3
Query: 18 LEVSEEDEDHLNRSAMELKYTGINFKSSETISLEVSEEDEDHLNQ 62
L V+ ED H+N + YTGIN+ S +I L+ EDH++Q
Sbjct: 1188 LHVAIEDNRHMNTLSKSGAYTGINY-ISFSIILQRLNTLEDHIHQ 1057
>TC222193 UP|Q39873 (Q39873) Lea protein precursor, complete
Length = 1748
Score = 30.4 bits (67), Expect = 1.1
Identities = 31/157 (19%), Positives = 68/157 (42%), Gaps = 3/157 (1%)
Frame = +3
Query: 11 ETSETISLEVSEEDEDHLNRSAMELKYTGINFKSSETISLEVSEEDEDHLNQSAMKLKET 70
+ S+ + +V++ +D+ +S Y G ++ + + + + + +D+ ++ K KE
Sbjct: 924 QKSKEYAGDVAQNAKDYAQKSK---DYAGDAVQNIKDYANDAAHKSKDYAGDASHKSKE- 1091
Query: 71 RINFKSSETSLEVSEEDKD---DIIELVIEDHEHALPQSAMVVKKTGINFKTSETSLKVS 127
+S+ + E +++ KD D + E E+A + + G + K S
Sbjct: 1092 -----ASDYASETAQKTKDYVGDAAQKSKEASEYASDAAQRTKEYAG-------DATKRS 1235
Query: 128 KEDEDHLNRSAMELKEAGISFKKSKTRSLKDVSFNRG 164
KE DH N A + K+ + L+D++ G
Sbjct: 1236 KEASDHANDMARKTKDYASDTAQRTKEKLQDIASEAG 1346
>TC221344
Length = 838
Score = 30.0 bits (66), Expect = 1.4
Identities = 22/110 (20%), Positives = 49/110 (44%), Gaps = 6/110 (5%)
Frame = +3
Query: 40 INFKSSETISLEVSEEDEDHLNQSAMKLKETRINFKSSETSLEVSEEDKDDIIELVIEDH 99
++ ++ +T + E E D + + + L+ R + + + S+ED+DD++ L D
Sbjct: 33 VSTEAGQTAACEAEECDSGNDDNQDVSLR--RSSNRKRRVVFDFSDEDEDDVVSLASPDL 206
Query: 100 EH------ALPQSAMVVKKTGINFKTSETSLKVSKEDEDHLNRSAMELKE 143
+ + +KT +NF E + KE+ ++ + L+E
Sbjct: 207 PNKQSSLDSRQNDKKTSEKTTLNFDLQEENKSGVKEERATEQKAHLPLRE 356
>TC226010 weakly similar to UP|Q8YPJ4 (Q8YPJ4) 50S ribosomal protein L18,
partial (78%)
Length = 860
Score = 29.6 bits (65), Expect = 1.9
Identities = 14/32 (43%), Positives = 21/32 (64%)
Frame = +1
Query: 154 RSLKDVSFNRGVLRLPKLVVDDHTECMFLNLI 185
RSL++ + RG RLP + VD HT +F +L+
Sbjct: 454 RSLQNPVWRRGSQRLPLIEVDTHTMAVFKHLL 549
>TC215566 similar to UP|Q9FSG4 (Q9FSG4) Wound-induced GSK-3-like protein
(Fragment), partial (62%)
Length = 1401
Score = 29.6 bits (65), Expect = 1.9
Identities = 23/82 (28%), Positives = 36/82 (43%)
Frame = +3
Query: 78 ETSLEVSEEDKDDIIELVIEDHEHALPQSAMVVKKTGINFKTSETSLKVSKEDEDHLNRS 137
E E ++K + IE + D E + S K N TS+ S V++ ++
Sbjct: 453 EPETEGKADEKTNTIETICTDQEQPIDTS-----KETSNVGTSDVST-VARTEKSGFEEL 614
Query: 138 AMELKEAGISFKKSKTRSLKDV 159
EL E I +KSK + KD+
Sbjct: 615 PKELNEMKIRDEKSKNNNEKDI 680
>TC208485 similar to UP|Q96424 (Q96424) O-methyltransferase, partial (68%)
Length = 830
Score = 27.7 bits (60), Expect = 7.1
Identities = 15/61 (24%), Positives = 29/61 (46%), Gaps = 13/61 (21%)
Frame = +1
Query: 193 DGTRKEVISFVCFI-------------DTIIDSAVDVAILIRSGIIINYLESDKAVAKLF 239
DGTR + SF F+ + ++D+ +D+ I + Y+E+D + ++F
Sbjct: 382 DGTRGYLASFTAFVCYPPLLQVWLNFKEAMVDADIDLFKKIHGVTMYQYMENDPKMNQIF 561
Query: 240 N 240
N
Sbjct: 562 N 564
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.132 0.358
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,863,399
Number of Sequences: 63676
Number of extensions: 123385
Number of successful extensions: 818
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 776
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 806
length of query: 315
length of database: 12,639,632
effective HSP length: 97
effective length of query: 218
effective length of database: 6,463,060
effective search space: 1408947080
effective search space used: 1408947080
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC148403.7


