

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148396.11 + phase: 0
(745 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
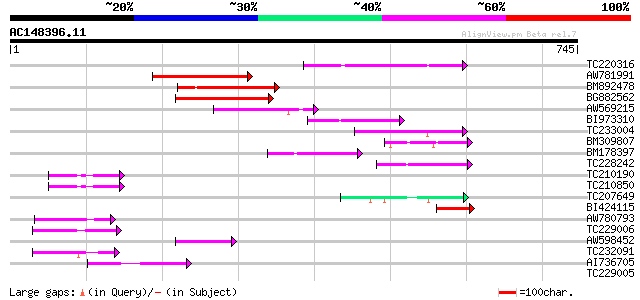
Score E
Sequences producing significant alignments: (bits) Value
TC220316 136 3e-32
AW781991 132 7e-31
BM892478 125 7e-29
BG882562 109 4e-24
AW569215 weakly similar to GP|4914326|gb| F14N23.12 {Arabidopsis... 92 1e-18
BI973310 80 3e-15
TC233004 weakly similar to PIR|G96565|G96565 F6D8.26 [imported] ... 67 4e-11
BM309807 similar to GP|5764395|gb| far-red impaired response pro... 66 6e-11
BM178397 similar to GP|4914326|gb| F14N23.12 {Arabidopsis thalia... 65 8e-11
TC228242 weakly similar to UP|Q9LIE5 (Q9LIE5) Far-red impaired r... 65 1e-10
TC210190 similar to UP|Q93Z68 (Q93Z68) AT3g59470/T16L24_20, part... 53 6e-07
TC210850 similar to UP|Q93Z68 (Q93Z68) AT3g59470/T16L24_20, part... 52 1e-06
TC207649 similar to UP|Q9SY66 (Q9SY66) F14N23.12 (At1g10240/F14N... 52 1e-06
BI424115 similar to GP|15982769|gb| At2g27110/T20P8.16 {Arabidop... 51 2e-06
AW780793 51 2e-06
TC229006 50 4e-06
AW598452 weakly similar to GP|15982769|gb| At2g27110/T20P8.16 {A... 47 4e-05
TC232091 47 4e-05
AI736705 similar to GP|11994736|db far-red impaired response pro... 44 3e-04
TC229005 36 0.071
>TC220316
Length = 988
Score = 136 bits (343), Expect = 3e-32
Identities = 80/217 (36%), Positives = 113/217 (51%), Gaps = 1/217 (0%)
Frame = +1
Query: 386 WNSESSEEFDLEWNNIISDYSLEGNGWLSTMYDLRSMWIPAYFKDTFMAGILRTTSRSES 445
+ S++ +E WN + Y L+ + WL MY R+ W+P Y K TF AGI +ES
Sbjct: 10 YQSQTVDEXXATWNXXXNKYGLKDDAWLKEMYQKRASWVPLYLKGTFFAGI----PMNES 177
Query: 446 ENSFFGNYLNHNLTLVEFWVRFDSALAAQRHKELFADNNTLHSNPELNMHMNLEKHAREV 505
+SFFG LN L+EF R++ L +R +E D NT + P L +E+ R +
Sbjct: 178 LDSFFGALLNAQTPLMEFIPRYERGLERRREEERKEDFNTSNFQPILQTKEPVEEQCRRL 357
Query: 506 YTHENFYIFQKELWSACVDCGIEGTKEEG-ENLSFSILDNAVRKHREVVYCLSNNIAHCS 564
YT F IFQKEL G + +E G N + KH V + SN CS
Sbjct: 358 YTLTVFKIFQKELLQCFSYLGFKIFEEGGLSRYMVRRCGNDMEKH-VVTFNASNISISCS 534
Query: 565 CKMFESEGIPCRHILFILKGKGFSEIPSHYIVNRWTK 601
C+MFE EG+ CRH+L + + E+PS YI++RWT+
Sbjct: 535 CQMFEYEGVLCRHVLRVFQILQLREVPSRYILHRWTR 645
>AW781991
Length = 419
Score = 132 bits (331), Expect = 7e-31
Identities = 60/132 (45%), Positives = 84/132 (63%)
Frame = +3
Query: 188 EQVGSYENIGCTQRDLQNYSRNLKELIKDSDADMFIDNFRRKREINPSFFYDYEADNEGK 247
++ G +G T+ D +NY RN + + D + +D R+ NP+FFY + D +
Sbjct: 24 KEYGGISKVGFTEVDCRNYMRNNRLRSLEGDIQLVLDYLRQMHAENPNFFYAVQGDEDQS 203
Query: 248 LKHVFWADGICRKNYSLFGDVVSFDTTYRTNKYFMIFAPFTGINHHRQSITFGAALLKNE 307
+ +VFWAD R NY+ FGD V+FDTTYR+N+Y + FA FTG+NHH Q + FG A L NE
Sbjct: 204 ITNVFWADPKARMNYTFFGDTVTFDTTYRSNRYRLPFAFFTGVNHHGQPVLFGCAFLINE 383
Query: 308 KEESFVWLFETF 319
E SFVWLF+T+
Sbjct: 384 SEASFVWLFKTW 419
>BM892478
Length = 429
Score = 125 bits (314), Expect = 7e-29
Identities = 59/134 (44%), Positives = 85/134 (63%)
Frame = +3
Query: 221 MFIDNFRRKREINPSFFYDYEADNEGKLKHVFWADGICRKNYSLFGDVVSFDTTYRTNKY 280
+ ++ F+ ++ + FFY E DN G ++FWADG R + S FGDV+ DT+YR Y
Sbjct: 27 ILLEYFQSRQAEDTGFFYAMEVDN-GNCMNIFWADGRSRYSCSHFGDVLVLDTSYRKTVY 203
Query: 281 FMIFAPFTGINHHRQSITFGAALLKNEKEESFVWLFETFLKAMGGHKPVMIITDQDGGMK 340
+ FA F G+NHH+Q + G AL+ +E EESF WLF+T+L+AM G P+ +I DQD ++
Sbjct: 204 LVPFATFVGVNHHKQPVLLGCALIADESEESFTWLFQTWLRAMSGRLPLTVIADQDIAIQ 383
Query: 341 NAIGAVLKGSSHRF 354
AI V + HRF
Sbjct: 384 RAIAKVFPVTHHRF 425
>BG882562
Length = 416
Score = 109 bits (273), Expect = 4e-24
Identities = 52/129 (40%), Positives = 80/129 (61%)
Frame = +3
Query: 218 DADMFIDNFRRKREINPSFFYDYEADNEGKLKHVFWADGICRKNYSLFGDVVSFDTTYRT 277
D ++ + F R + +NP+FFY + +++G+L++VFW D R YS FGDVV+FD+T +
Sbjct: 30 DPELISNYFCRIQLMNPNFFYVMDLNDDGQLRNVFWIDSRSRAAYSYFGDVVAFDSTCLS 209
Query: 278 NKYFMIFAPFTGINHHRQSITFGAALLKNEKEESFVWLFETFLKAMGGHKPVMIITDQDG 337
N Y + F G+NHH +S+ G LL +E E+++WLF +L M G P IIT+
Sbjct: 210 NNYEIPLVAFVGVNHHGKSVLLGCGLLADETFETYIWLFRAWLTCMTGRPPQTIITNXCK 389
Query: 338 GMKNAIGAV 346
M++AI V
Sbjct: 390 AMQSAIAEV 416
>AW569215 weakly similar to GP|4914326|gb| F14N23.12 {Arabidopsis thaliana},
partial (19%)
Length = 425
Score = 91.7 bits (226), Expect = 1e-18
Identities = 50/141 (35%), Positives = 74/141 (52%), Gaps = 4/141 (2%)
Frame = +1
Query: 269 VSFDTTYRTNKYFMIFAPFTGINHHRQSITFGAALLKNEKEESFVWLFETFLKAMGGHKP 328
V FDTTYR Y M+ + G++++ + F ALL++E +SF W + FL M G P
Sbjct: 10 VVFDTTYRVEAYDMLLGIWLGVDNNGMTCFFSCALLRDENIQSFSWALKAFLGFMKGKAP 189
Query: 329 VMIITDQDGGMKNAIGAVLKGSSHRFCMWHILKKLSE----KVGSSMDENSGFNDRFKSC 384
I+TD + +K AI L + H FC+WHIL K S+ +GS DE R
Sbjct: 190 QTILTDHNMWLKEAIAVELPQTKHAFCIWHILSKFSDWFSLLLGSQYDEWKAEFHR---- 357
Query: 385 VWNSESSEEFDLEWNNIISDY 405
++N E E+F+ W ++ Y
Sbjct: 358 LYNLEQVEDFEEGWRQMVDQY 420
>BI973310
Length = 457
Score = 80.1 bits (196), Expect = 3e-15
Identities = 41/127 (32%), Positives = 72/127 (56%)
Frame = +3
Query: 392 EEFDLEWNNIISDYSLEGNGWLSTMYDLRSMWIPAYFKDTFMAGILRTTSRSESENSFFG 451
+EF+ W++++ + + N WL ++Y+ R W+P Y ++TF G + +E SFF
Sbjct: 9 DEFESYWHSLLERFYVMDNEWLQSIYNSRQHWVPVYLRETFF-GEISLNEGNEYLISFFD 185
Query: 452 NYLNHNLTLVEFWVRFDSALAAQRHKELFADNNTLHSNPELNMHMNLEKHAREVYTHENF 511
Y+ + TL +++ A+++ KEL AD +T +S+P L +EK A +YT + F
Sbjct: 186 GYVFSSTTLQVLVRQYEKAVSSWHEKELKADYDTTNSSPVLKTPSPMEKQAASLYTRKIF 365
Query: 512 YIFQKEL 518
FQ+EL
Sbjct: 366 MKFQEEL 386
>TC233004 weakly similar to PIR|G96565|G96565 F6D8.26 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(8%)
Length = 551
Score = 66.6 bits (161), Expect = 4e-11
Identities = 44/156 (28%), Positives = 68/156 (43%), Gaps = 7/156 (4%)
Frame = +1
Query: 453 YLNHNLTLVEFWVRFDSALAAQRHKELFADNNTLHSNPELNMHMNLEKHAREVYTHENFY 512
Y++ + + EF ++D L + KE AD T + + EL N E +VYT E F
Sbjct: 7 YVHKHTSFKEFVDKYDLVLHRKHLKEAMADLETRNVSFELKTRCNFEVQLAKVYTKEIFQ 186
Query: 513 IFQKELWSACVDCGIEGTKEEGENLSFSILDNAVR-------KHREVVYCLSNNIAHCSC 565
FQ E+ G +++ + + K EV+Y + C C
Sbjct: 187 KFQSEVEGMYSCFNTRQVSVNGSIITYVVKERVEVEGNEKGVKSFEVLYETTELDIRCIC 366
Query: 566 KMFESEGIPCRHILFILKGKGFSEIPSHYIVNRWTK 601
+F +G CR L +L G EIPS YI++RW +
Sbjct: 367 SLFNYKGYLCRRALNVLNYNGIEEIPSRYILHRWRR 474
>BM309807 similar to GP|5764395|gb| far-red impaired response protein
{Arabidopsis thaliana}, partial (15%)
Length = 428
Score = 65.9 bits (159), Expect = 6e-11
Identities = 43/123 (34%), Positives = 61/123 (48%), Gaps = 7/123 (5%)
Frame = +3
Query: 493 NMHMNL----EKHAREVYTHENFYIFQKELWSACVDCGIEGTKEEGENLSFSILDNAVRK 548
+ H+NL EK VYTH F FQ E+ G + E G+ + K
Sbjct: 18 SQHLNLLHLXEKQMSTVYTHAIFKKFQVEVLGVA---GCQSRIEAGDGTIAKFIVQDYEK 188
Query: 549 HREVVYC---LSNNIAHCSCKMFESEGIPCRHILFILKGKGFSEIPSHYIVNRWTKLATS 605
E + LS+ ++ C C++FE +G CRH L +L+ G S +PSHYI+ RWTK A
Sbjct: 189 DEEFLVTWNELSSEVS-CFCRLFEYKGFLCRHALSVLQRCGCSCVPSHYILKRWTKDAKI 365
Query: 606 KPI 608
K +
Sbjct: 366 KEL 374
>BM178397 similar to GP|4914326|gb| F14N23.12 {Arabidopsis thaliana}, partial
(20%)
Length = 422
Score = 65.5 bits (158), Expect = 8e-11
Identities = 36/127 (28%), Positives = 63/127 (49%), Gaps = 2/127 (1%)
Frame = +1
Query: 339 MKNAIGAVLKGSSHRFCMWHILKKLSEKVGSSMDENSGFNDRFKSC--VWNSESSEEFDL 396
+K A+ + + H FC+W I+ K + + E +ND ++N ES E+F+L
Sbjct: 13 LKEALSTEMPTTKHAFCIWMIVAKFPSWFNAVLGER--YNDWKAEFYRLYNLESVEDFEL 186
Query: 397 EWNNIISDYSLEGNGWLSTMYDLRSMWIPAYFKDTFMAGILRTTSRSESENSFFGNYLNH 456
W + + L N + +Y RS+W + + F+AG + TT +S+S N+F +L+
Sbjct: 187 GWREMACSFGLHSNRHMVNLYSSRSLWALPFLRSHFLAG-MTTTGQSKSINAFIQRFLSA 363
Query: 457 NLTLVEF 463
L F
Sbjct: 364 QTRLAHF 384
>TC228242 weakly similar to UP|Q9LIE5 (Q9LIE5) Far-red impaired response
protein; Mutator-like transposase-like protein;
phytochrome A signaling protein-like, partial (25%)
Length = 1354
Score = 65.1 bits (157), Expect = 1e-10
Identities = 41/127 (32%), Positives = 60/127 (46%)
Frame = +2
Query: 482 DNNTLHSNPELNMHMNLEKHAREVYTHENFYIFQKELWSACVDCGIEGTKEEGENLSFSI 541
D++T + L LEK ++TH F Q E+ A V C + +++ + +
Sbjct: 2 DSDTWNKVATLKTPSPLEKSVAGIFTHAVFKKIQAEVIGA-VACHPKADRQDDTTIVHRV 178
Query: 542 LDNAVRKHREVVYCLSNNIAHCSCKMFESEGIPCRHILFILKGKGFSEIPSHYIVNRWTK 601
D K VV + C C++FE G CRH LF+L+ G S PS YI+ RWTK
Sbjct: 179 HDMETNKDFFVVVNQVKSELSCICRLFEYRGYLCRHALFVLQYSGQSVFPSQYILKRWTK 358
Query: 602 LATSKPI 608
A + I
Sbjct: 359 DAKVRNI 379
>TC210190 similar to UP|Q93Z68 (Q93Z68) AT3g59470/T16L24_20, partial (65%)
Length = 866
Score = 52.8 bits (125), Expect = 6e-07
Identities = 35/102 (34%), Positives = 56/102 (54%), Gaps = 2/102 (1%)
Frame = +1
Query: 52 FYKAYAYVAGFSVRNS-IKTKDKDGVK-WKYFLCSKEGFKEEKKVDKPQLLIAENSLSKS 109
FY AYA GF VR S + +DG + +C+KEGF+ DK + ++
Sbjct: 34 FYNAYATEVGFIVRVSKLSRSRRDGTAIGRTLVCNKEGFR---MADKREKIV-------- 180
Query: 110 RKRKLTREGCKARLVLKRTIDGKYEVSNFYEGHSHGLVSPSK 151
R+R TR GC+A +++++ GK+ V+ F + H+H L +P K
Sbjct: 181 RQRAETRVGCRAMIMVRKLSSGKWVVAKFVKEHTHPL-TPGK 303
>TC210850 similar to UP|Q93Z68 (Q93Z68) AT3g59470/T16L24_20, partial (63%)
Length = 1326
Score = 52.0 bits (123), Expect = 1e-06
Identities = 34/102 (33%), Positives = 56/102 (54%), Gaps = 2/102 (1%)
Frame = +1
Query: 52 FYKAYAYVAGFSVRNS-IKTKDKDGVK-WKYFLCSKEGFKEEKKVDKPQLLIAENSLSKS 109
FY AYA GF VR S + +DG + +C+KEGF+ DK + ++
Sbjct: 16 FYNAYATDVGFIVRVSKLSRSRRDGTAIGRTLVCNKEGFR---MADKREKIV-------- 162
Query: 110 RKRKLTREGCKARLVLKRTIDGKYEVSNFYEGHSHGLVSPSK 151
R+R TR GC+A +++++ GK+ ++ F + H+H L +P K
Sbjct: 163 RQRAETRVGCRAMIMVRKLSSGKWVIAKFVKEHTHPL-TPGK 285
>TC207649 similar to UP|Q9SY66 (Q9SY66) F14N23.12 (At1g10240/F14N23_12),
partial (30%)
Length = 971
Score = 51.6 bits (122), Expect = 1e-06
Identities = 50/188 (26%), Positives = 75/188 (39%), Gaps = 20/188 (10%)
Frame = +2
Query: 435 GILRTTSRSESENSFFGNYLNHNL--TLVEFWVRFDSALA--------AQRHKELFADNN 484
G L S+++ F+ L HN L++F++ + LA A K+
Sbjct: 23 GALPKKSKAKRSRRFYMPDLIHNK*QLLIQFFLSAQTRLAHFVEQVAVAVDFKDQTG*QQ 202
Query: 485 TLHSNPE---LNMHMNLEKHAREVYTHENFYIFQKELWSACVDCGIEGTKEEGENLSFSI 541
T+ N + L +E HA + T F Q++L A SFSI
Sbjct: 203 TMQQNLQNVCLKTGAPMESHAATILTPFAFSKLQEQLVLAA------------HYASFSI 346
Query: 542 LDNAVRKH-------REVVYCLSNNIAHCSCKMFESEGIPCRHILFILKGKGFSEIPSHY 594
D + +H R+V + I CSC FE GI CRH L +L +IP Y
Sbjct: 347 EDGFLVRHHTKAEGGRKVYWAPQEGIISCSCHQFEFTGILCRHSLRVLSTGNCFQIPDRY 526
Query: 595 IVNRWTKL 602
+ RW ++
Sbjct: 527 LPIRWRRI 550
>BI424115 similar to GP|15982769|gb| At2g27110/T20P8.16 {Arabidopsis
thaliana}, partial (9%)
Length = 402
Score = 50.8 bits (120), Expect = 2e-06
Identities = 22/50 (44%), Positives = 30/50 (60%)
Frame = +2
Query: 561 AHCSCKMFESEGIPCRHILFILKGKGFSEIPSHYIVNRWTKLATSKPIFD 610
A+CSC+MFE GI C+HIL + +P HYI+ RWT+ A + D
Sbjct: 83 ANCSCQMFEYAGILCKHILTVFTVTNVLTLPPHYILKRWTRNAKNSAGLD 232
>AW780793
Length = 419
Score = 50.8 bits (120), Expect = 2e-06
Identities = 33/108 (30%), Positives = 58/108 (53%), Gaps = 2/108 (1%)
Frame = +3
Query: 33 DELKPVIGKVFDTLVEGGDFYKAYAYVAGFSVR--NSIKTKDKDGVKWKYFLCSKEGFKE 90
+ ++P IG F++ E +FY AY GF+VR ++ +++ + V + F+CSKEGF+
Sbjct: 120 EAVEPFIGMEFNSREEAREFYIAYGRRIGFTVRIHHNRRSRVNNQVIGQDFVCSKEGFRA 299
Query: 91 EKKVDKPQLLIAENSLSKSRKRKLTREGCKARLVLKRTIDGKYEVSNF 138
+K + + ++ TREGC+A + L GK+ V+ F
Sbjct: 300 KKYLHRKDRVLP--------PPPATREGCQAMIRLALRDRGKWVVTKF 419
>TC229006
Length = 991
Score = 50.1 bits (118), Expect = 4e-06
Identities = 38/121 (31%), Positives = 58/121 (47%), Gaps = 4/121 (3%)
Frame = +2
Query: 31 CEDEL--KPVIGKVFDTLVEGGDFYKAYAYVAGFSVR-NSIKTKDKDG-VKWKYFLCSKE 86
CED +P G F++ FY YA GF +R S + ++DG + + C+KE
Sbjct: 77 CEDHAIQEPYEGMEFESEDAAKIFYDEYARRLGFVMRVMSCRRPERDGRILARRLGCNKE 256
Query: 87 GFKEEKKVDKPQLLIAENSLSKSRKRKLTREGCKARLVLKRTIDGKYEVSNFYEGHSHGL 146
G+ + I S + R TREGCKA + +K GK+ ++ F + H+H L
Sbjct: 257 GYC---------VSIRGKFASVRKPRASTREGCKAMIHIKYNKSGKWVITKFVKDHNHPL 409
Query: 147 V 147
V
Sbjct: 410 V 412
>AW598452 weakly similar to GP|15982769|gb| At2g27110/T20P8.16 {Arabidopsis
thaliana}, partial (7%)
Length = 256
Score = 46.6 bits (109), Expect = 4e-05
Identities = 24/80 (30%), Positives = 40/80 (50%)
Frame = +2
Query: 218 DADMFIDNFRRKREINPSFFYDYEADNEGKLKHVFWADGICRKNYSLFGDVVSFDTTYRT 277
DA ++ +R + N FY + D + ++ +V A R +Y GD S D Y
Sbjct: 17 DAHNLLEYCKRMQAENHGCFYAIQLDEDDRVSNVL*AGTRSRTSYKHMGDAESLDAAYGK 196
Query: 278 NKYFMIFAPFTGINHHRQSI 297
++Y + +AP+TG N H + I
Sbjct: 197 HRYAVSYAPYTGRNCHGRMI 256
>TC232091
Length = 530
Score = 46.6 bits (109), Expect = 4e-05
Identities = 37/124 (29%), Positives = 57/124 (45%), Gaps = 9/124 (7%)
Frame = +1
Query: 30 ACEDEL--KPVIGKVFDTLVEGGDFYKAYAYVAGFSVR--NSIKTKDKDGVKWKYFLCSK 85
ACE +P +G F + +FY+ YA GF VR +++ + + F C+K
Sbjct: 109 ACEGSSIEEPNVGMEFQSEEAAKNFYEEYARREGFVVRLDRCHRSEVDKQIISRRFSCNK 288
Query: 86 EGF-----KEEKKVDKPQLLIAENSLSKSRKRKLTREGCKARLVLKRTIDGKYEVSNFYE 140
+GF + K V KP+ I REGC+A + +K GK+ V+ F +
Sbjct: 289 QGFHVRVRNKTKPVHKPRASI--------------REGCEAMMYVKVNTCGKWVVTKFVK 426
Query: 141 GHSH 144
HSH
Sbjct: 427 EHSH 438
>AI736705 similar to GP|11994736|db far-red impaired response protein;
Mutator-like transposase-like protein; phytochrome A
signaling, partial (14%)
Length = 383
Score = 43.9 bits (102), Expect = 3e-04
Identities = 26/136 (19%), Positives = 58/136 (42%)
Frame = +3
Query: 103 ENSLSKSRKRKLTREGCKARLVLKRTIDGKYEVSNFYEGHSHGLVSPSKRQFLRSARNVT 162
++S + + +R ++ CKA + +KR DGK+ + +F + H+H L+
Sbjct: 39 QDSENSTGRRSCSKTDCKASMHVKRRSDGKWVIHSFVKEHNHELL--------------- 173
Query: 163 SVHKNILFSCNRANVGTSKSYQIMKEQVGSYENIGCTQRDLQNYSRNLKELIKDSDADMF 222
+ T + Y M Q Y+ + + + + + ++ +A +
Sbjct: 174 --------PAQAVSEQTRRMYAAMARQFAEYKTVVGLKNEKNPFDKGRNLGLESGEAKLM 329
Query: 223 IDNFRRKREINPSFFY 238
+D F + + +N +FFY
Sbjct: 330 LDFFIQMQNMNSNFFY 377
>TC229005
Length = 679
Score = 35.8 bits (81), Expect = 0.071
Identities = 15/33 (45%), Positives = 22/33 (66%)
Frame = +1
Query: 115 TREGCKARLVLKRTIDGKYEVSNFYEGHSHGLV 147
TREGCKA + +K GK+ ++ F + H+H LV
Sbjct: 7 TREGCKAMIHIKYDKSGKWVIAKFVKDHNHPLV 105
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.134 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 35,878,771
Number of Sequences: 63676
Number of extensions: 523831
Number of successful extensions: 2551
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 2523
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2542
length of query: 745
length of database: 12,639,632
effective HSP length: 104
effective length of query: 641
effective length of database: 6,017,328
effective search space: 3857107248
effective search space used: 3857107248
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC148396.11


