

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148396.10 + phase: 0 /partial
(673 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
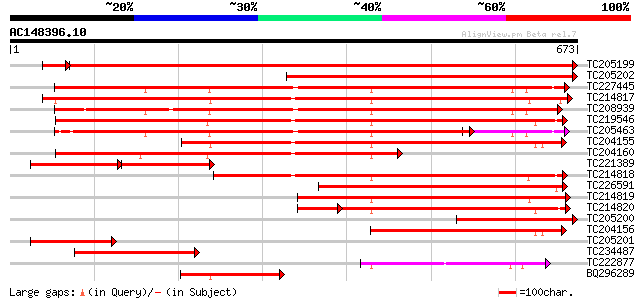
Score E
Sequences producing significant alignments: (bits) Value
TC205199 homologue to UP|HS7M_PEA (P37900) Heat shock 70 kDa pro... 1077 0.0
TC205202 homologue to UP|HS7M_PEA (P37900) Heat shock 70 kDa pro... 612 e-175
TC227445 homologue to UP|O22639 (O22639) Endoplasmic reticulum H... 582 e-166
TC214817 homologue to UP|Q9M4E7 (Q9M4E7) Heat shock protein 70, ... 561 e-160
TC208939 UP|Q39830 (Q39830) BiP isoform A, complete 556 e-158
TC219546 UP|HS70_SOYBN (P26413) Heat shock 70 kDa protein, complete 555 e-158
TC205463 UP|Q39804 (Q39804) BiP isoform B, complete 502 e-142
TC204155 homologue to UP|Q84QJ3 (Q84QJ3) Heat shock protein 70, ... 413 e-115
TC204160 homologue to UP|HS72_LYCES (P27322) Heat shock cognate ... 400 e-112
TC221389 homologue to UP|HS7M_PHAVU (Q01899) Heat shock 70 kDa p... 202 e-104
TC214818 homologue to UP|Q8GSN4 (Q8GSN4) Non-cell-autonomous hea... 366 e-101
TC226591 homologue to PRF|1909352A.0|445605|1909352A heat shock ... 321 6e-88
TC214819 homologue to UP|Q8GSN2 (Q8GSN2) Cell-autonomous heat sh... 283 2e-76
TC214820 homologue to UP|Q9M6R1 (Q9M6R1) High molecular weight h... 235 4e-71
TC205200 similar to UP|HS7M_PEA (P37900) Heat shock 70 kDa prote... 251 1e-66
TC204156 homologue to UP|Q40151 (Q40151) Hsc70 protein, partial ... 205 6e-53
TC205201 similar to UP|HS7M_PEA (P37900) Heat shock 70 kDa prote... 178 8e-45
TC234487 similar to UP|Q9SAU8 (Q9SAU8) HSP70, partial (23%) 152 4e-37
TC222877 weakly similar to UP|Q94614 (Q94614) Heat shock 70kDa p... 151 8e-37
BQ296289 homologue to PIR|T45517|T45 heat shock protein 70 cyto... 149 4e-36
>TC205199 homologue to UP|HS7M_PEA (P37900) Heat shock 70 kDa protein,
mitochondrial precursor, partial (94%)
Length = 2204
Score = 1077 bits (2785), Expect(2) = 0.0
Identities = 555/603 (92%), Positives = 581/603 (96%), Gaps = 1/603 (0%)
Frame = +3
Query: 72 KNPKVVENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNPENTISGAKRLIGRRFDDPQ 131
+NPKV+ENSEGARTTPSVVAF QK ELLVGTPAKRQAVTNP NT+ G KRLIGRRFDD Q
Sbjct: 177 QNPKVIENSEGARTTPSVVAFNQKAELLVGTPAKRQAVTNPTNTLFGTKRLIGRRFDDSQ 356
Query: 132 TQKEMKMVPYKIVKAPNGDAWVEAKGQQYSPSQIGAFVLTKMKETAEAYLGKTVPKAVIT 191
TQKEMKMVPYKIVKAPNGDAWVEA GQQYSPSQ+GAFVLTKMKETAE+YLGK+V KAVIT
Sbjct: 357 TQKEMKMVPYKIVKAPNGDAWVEANGQQYSPSQVGAFVLTKMKETAESYLGKSVSKAVIT 536
Query: 192 VPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALSYGMN-KEGLIAVFDLGGGTFDVS 250
VPAYFNDAQRQATKDAGRIAGL+V RIINEPTAAALSYGMN KEGLIAVFDLGGGTFDVS
Sbjct: 537 VPAYFNDAQRQATKDAGRIAGLDVQRIINEPTAAALSYGMNNKEGLIAVFDLGGGTFDVS 716
Query: 251 ILEISNGVFEVKATNGDTFLGGEDFDNALLDFLVNEFKRSESIDLSKDKLALQRLREAAE 310
ILEISNGVFEVKATNGDTFLGGEDFDNALLDFLVNEFKR+E+IDLSKDKLALQRLREAAE
Sbjct: 717 ILEISNGVFEVKATNGDTFLGGEDFDNALLDFLVNEFKRTENIDLSKDKLALQRLREAAE 896
Query: 311 KAKIELSSTSQTEINLPFITADASGAKHLNITLTRSKFEALVNNLIERTKAPCQSCLKDA 370
KAKIELSSTSQTEINLPFITADASGAKHLNITLTRSKFEALVN+LIERTKAPC+SCLKDA
Sbjct: 897 KAKIELSSTSQTEINLPFITADASGAKHLNITLTRSKFEALVNHLIERTKAPCKSCLKDA 1076
Query: 371 NISTKDIDEVLLVGGMTRVPKVQEVVSTIFGKSPCKGVNPDEAVAMGAALQGGILRGDVK 430
N+S K++DEVLLVGGMTRVPKVQEVVS IFGKSP KGVNPDEAVAMGAA+QGGILRGDVK
Sbjct: 1077NVSIKEVDEVLLVGGMTRVPKVQEVVSAIFGKSPSKGVNPDEAVAMGAAIQGGILRGDVK 1256
Query: 431 ELLLLDVTPLSLGIETLGGIFTRLINRNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREM 490
ELLLLDVTPLSLGIETLGGIFTRLINRNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREM
Sbjct: 1257ELLLLDVTPLSLGIETLGGIFTRLINRNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREM 1436
Query: 491 ASDNKMLGEFELVGLPPAPRGMPQIEVTFDIDANGIVTVSAKDKSTGKEQQITIKSSGGL 550
A DNK LGEFELVG+PPAPRGMPQIEVTFDIDANGIVTVSAKDKSTGKEQQITI+SSGGL
Sbjct: 1437AVDNKSLGEFELVGIPPAPRGMPQIEVTFDIDANGIVTVSAKDKSTGKEQQITIRSSGGL 1616
Query: 551 SDDEIQNMVKEAELHAQKDQERKSLIDIRNSADTSIYSIEKSLSEYREKIPAEVAKEIED 610
S+DEI MVKEAELHAQKDQERK+LIDIRNSADTSIYSIEKSL EYR+KIP+EVAKEIED
Sbjct: 1617SEDEIDKMVKEAELHAQKDQERKALIDIRNSADTSIYSIEKSLGEYRDKIPSEVAKEIED 1796
Query: 611 AVSDLRSAMAGDSADEIKSKLDAANKAVSKIGEHMSGGSSSGPSSDGSQGGDQAPEAEYE 670
AVSDLR+AMAGD+ADEIK+KLDAANKAVSKIGEHMSGGSS S+ GSQGG+QAPEAEYE
Sbjct: 1797AVSDLRTAMAGDNADEIKAKLDAANKAVSKIGEHMSGGSSGSSSAGGSQGGEQAPEAEYE 1976
Query: 671 EVK 673
EVK
Sbjct: 1977EVK 1985
Score = 65.5 bits (158), Expect(2) = 0.0
Identities = 31/34 (91%), Positives = 33/34 (96%)
Frame = +1
Query: 39 SSLSRPFSSRAAGNDVIGIDLGTTNSCVSVMEGK 72
+SLSRPFSS+ AGNDVIGIDLGTTNSCVSVMEGK
Sbjct: 1 ASLSRPFSSKPAGNDVIGIDLGTTNSCVSVMEGK 102
>TC205202 homologue to UP|HS7M_PEA (P37900) Heat shock 70 kDa protein,
mitochondrial precursor, partial (51%)
Length = 1228
Score = 612 bits (1578), Expect = e-175
Identities = 313/345 (90%), Positives = 332/345 (95%)
Frame = +2
Query: 329 ITADASGAKHLNITLTRSKFEALVNNLIERTKAPCQSCLKDANISTKDIDEVLLVGGMTR 388
+ ADASGAKHLNITLTRSKFEALVN+LIERTK PC+SCLKDANIS KD+DEVLLVGGMTR
Sbjct: 2 VGADASGAKHLNITLTRSKFEALVNHLIERTKVPCKSCLKDANISIKDVDEVLLVGGMTR 181
Query: 389 VPKVQEVVSTIFGKSPCKGVNPDEAVAMGAALQGGILRGDVKELLLLDVTPLSLGIETLG 448
VPKVQEVVS IFGKSP KGVNPDEAVAMGAA+QGGILRGDVKELLLLDVTPLSLGIETLG
Sbjct: 182 VPKVQEVVSEIFGKSPSKGVNPDEAVAMGAAIQGGILRGDVKELLLLDVTPLSLGIETLG 361
Query: 449 GIFTRLINRNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGEFELVGLPPA 508
GIFTRLINRNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMA+DNKMLGEF+LVG+PPA
Sbjct: 362 GIFTRLINRNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMAADNKMLGEFDLVGIPPA 541
Query: 509 PRGMPQIEVTFDIDANGIVTVSAKDKSTGKEQQITIKSSGGLSDDEIQNMVKEAELHAQK 568
PRG+PQIEVTFDIDANGIVTVSAKDKSTGKEQQITI+SSGGLS+DEI+ MVKEAELHAQK
Sbjct: 542 PRGLPQIEVTFDIDANGIVTVSAKDKSTGKEQQITIRSSGGLSEDEIEKMVKEAELHAQK 721
Query: 569 DQERKSLIDIRNSADTSIYSIEKSLSEYREKIPAEVAKEIEDAVSDLRSAMAGDSADEIK 628
DQERK+LIDIRNSADT+IYSIEKSL EYR+KIP+EVAKEIEDAVSDLR AM+ D+ DEIK
Sbjct: 722 DQERKALIDIRNSADTTIYSIEKSLGEYRDKIPSEVAKEIEDAVSDLRKAMSEDNVDEIK 901
Query: 629 SKLDAANKAVSKIGEHMSGGSSSGPSSDGSQGGDQAPEAEYEEVK 673
SKLDAANKAVSKIGEHMSGGSS G S+ GSQGGDQAPEAEYEEVK
Sbjct: 902 SKLDAANKAVSKIGEHMSGGSSGGSSAGGSQGGDQAPEAEYEEVK 1036
>TC227445 homologue to UP|O22639 (O22639) Endoplasmic reticulum HSC70-cognate
binding protein precursor, complete
Length = 2254
Score = 582 bits (1501), Expect = e-166
Identities = 320/635 (50%), Positives = 430/635 (67%), Gaps = 24/635 (3%)
Frame = +2
Query: 54 VIGIDLGTTNSCVSVMEGKNPKVVENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNPE 113
VIGIDLGTT SCV V + + +++ N +G R TPS VAFT E L+G AK A NPE
Sbjct: 215 VIGIDLGTTYSCVGVYKNGHVEIIANDQGNRITPSWVAFTDS-ERLIGEAAKNLAAVNPE 391
Query: 114 NTISGAKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEAKGQQ-----YSPSQIGAF 168
TI KRLIGR+F+D + Q++MK+VPYKIV +G +++ K + +SP +I A
Sbjct: 392 RTIFDVKRLIGRKFEDKEVQRDMKLVPYKIVNK-DGKPYIQVKIKDGETKVFSPEEISAM 568
Query: 169 VLTKMKETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALS 228
+LTKMKETAEA+LGK + AV+TVPAYFNDAQRQATKDAG IAGL V RIINEPTAAA++
Sbjct: 569 ILTKMKETAEAFLGKKINDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAAAIA 748
Query: 229 YGMNKEG---LIAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLVN 285
YG++K+G I VFDLGGGTFDVSIL I NGVFEV ATNGDT LGGEDFD ++++ +
Sbjct: 749 YGLDKKGGEKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTHLGGEDFDQRIMEYFIK 928
Query: 286 EFKRSESIDLSKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLTR 345
K+ D+SKD AL +LR AE+AK LSS Q + + + ++ LTR
Sbjct: 929 LIKKKHGKDISKDNRALGKLRREAERAKRALSSQHQVRVEIESLFDGVDFSE----PLTR 1096
Query: 346 SKFEALVNNLIERTKAPCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIF-GKSP 404
++FE L N+L +T P + ++DA + IDE++LVGG TR+PKVQ+++ F GK P
Sbjct: 1097ARFEELNNDLFRKTMGPVKKAMEDAGLQKSQIDEIVLVGGSTRIPKVQQLLKDYFDGKEP 1276
Query: 405 CKGVNPDEAVAMGAALQGGILRGD----VKELLLLDVTPLSLGIETLGGIFTRLINRNTT 460
KGVNPDEAVA GAA+QG IL G+ K++LLLDV PL+LGIET+GG+ T+LI RNT
Sbjct: 1277NKGVNPDEAVAYGAAVQGSILSGEGGEETKDILLLDVAPLTLGIETVGGVMTKLIPRNTV 1456
Query: 461 IPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQIEVTFD 520
IPTKKSQVF+T D QT V I+V +GER + D ++LG+F+L G+PPAPRG PQIEVTF+
Sbjct: 1457IPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGKFDLSGIPPAPRGTPQIEVTFE 1636
Query: 521 IDANGIVTVSAKDKSTGKEQQITI-KSSGGLSDDEIQNMVKEAELHAQKDQERKSLIDIR 579
+DANGI+ V A+DK TGK ++ITI G LS +EI MV+EAE A++D++ K ID R
Sbjct: 1637VDANGILNVKAEDKGTGKSEKITITNEKGRLSQEEIDRMVREAEEFAEEDKKVKERIDAR 1816
Query: 580 NSADTSIYSIEKSLSE---YREKIPAEVAKEIEDAV-------SDLRSAMAGDSADEIKS 629
NS +T +Y+++ +S+ +K+ ++ ++IE AV D +S D +++K
Sbjct: 1817NSLETYVYNMKNQVSDKDKLADKLESDEKEKIETAVKEALEWLDDNQSVEKEDYEEKLKE 1996
Query: 630 KLDAANKAVSKIGEHMSGGSSSGPSSDGSQGGDQA 664
N +S + + SGG+ G + G + D +
Sbjct: 1997VEAVCNPIISAVYQR-SGGAPGGGGASGEEDKDDS 2098
>TC214817 homologue to UP|Q9M4E7 (Q9M4E7) Heat shock protein 70, complete
Length = 2364
Score = 561 bits (1446), Expect = e-160
Identities = 319/668 (47%), Positives = 429/668 (63%), Gaps = 38/668 (5%)
Frame = +2
Query: 39 SSLSRPFSSRAAGND---VIGIDLGTTNSCVSVMEGKNPKVVENSEGARTTPSVVAFTQK 95
SS S P + AG IGIDLGTT SCV V + +++ N +G RTTPS V FT
Sbjct: 56 SSFSTPHAETMAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVGFTDT 235
Query: 96 GELLVGTPAKRQAVTNPENTISGAKRLIGRRFDDPQTQKEMKMVPYKIVK--APNGDAWV 153
E L+G AK Q NP NT+ AKRLIGRRF D Q ++K+ P+K++ A V
Sbjct: 236 -ERLIGDAAKNQVAMNPINTVFDAKRLIGRRFSDSSVQSDIKLWPFKVIPGAADKPMIVV 412
Query: 154 EAKGQ--QYSPSQIGAFVLTKMKETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIA 211
KG+ Q++ +I + VL KM+E AEAYLG TV AV+TVPAYFND+QRQATKDAG IA
Sbjct: 413 NYKGEEKQFAAEEISSMVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIA 592
Query: 212 GLEVLRIINEPTAAALSYGMNKEGL------IAVFDLGGGTFDVSILEISNGVFEVKATN 265
GL V+RIINEPTAAA++YG++K+ + +FDLGGGTFDVS+L I G+FEVKAT
Sbjct: 593 GLNVMRIINEPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATA 772
Query: 266 GDTFLGGEDFDNALLDFLVNEFKRSESIDLSKDKLALQRLREAAEKAKIELSSTSQTEIN 325
GDT LGGEDFDN +++ V EFKR D+S + AL+RLR A E+AK LSST+QT I
Sbjct: 773 GDTHLGGEDFDNRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIE 952
Query: 326 LPFITADASGAKHLNITLTRSKFEALVNNLIERTKAPCQSCLKDANISTKDIDEVLLVGG 385
+ + T+TR++FE L +L + P + CL+DA + + +D+V+LVGG
Sbjct: 953 IDSLYEGID----FYSTVTRARFEELNMDLFRKCMEPVEKCLRDAKMDKRSVDDVVLVGG 1120
Query: 386 MTRVPKVQEVVSTIF-GKSPCKGVNPDEAVAMGAALQGGILRGD----VKELLLLDVTPL 440
TR+PKVQ+++ F GK CK +NPDEAVA GAA+Q IL G+ V++LLLLDVTPL
Sbjct: 1121STRIPKVQQLLQDFFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPL 1300
Query: 441 SLGIETLGGIFTRLINRNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGEF 500
SLG+ET GG+ T LI RNTTIPTKK QVFST +DNQ V I+V +GER DN +LG+F
Sbjct: 1301SLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVFEGERARTRDNNLLGKF 1480
Query: 501 ELVGLPPAPRGMPQIEVTFDIDANGIVTVSAKDKSTGKEQQITI-KSSGGLSDDEIQNMV 559
EL G+PPAPRG+PQI V FDIDANGI+ VSA+DK+TG++ +ITI G LS ++I+ MV
Sbjct: 1481ELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEDIEKMV 1660
Query: 560 KEAELHAQKDQERKSLIDIRNSADTSIYSIEKSLSEYR--EKIPAEVAKEIEDAVSDL-- 615
+EAE + +D+E K ++ +N+ + Y++ ++ + + EK+ K+IEDA+
Sbjct: 1661QEAEKYKSEDEEHKKKVEAKNALENYAYNMRNTVKDDKIGEKLDPADKKKIEDAIEQAIQ 1840
Query: 616 -----RSAMAGDSADEIKSKLDAANKAVSKIGEHMSGGSSSG----------PSSDGSQG 660
+ A A + D++K N ++K+ + +G G P S GS
Sbjct: 1841WLDSNQLAEADEFEDKMKELESICNPIIAKMYQGGAGPDVGGAGAAEDEYAAPPSGGSGA 2020
Query: 661 GDQAPEAE 668
G + E +
Sbjct: 2021GPKIEEVD 2044
>TC208939 UP|Q39830 (Q39830) BiP isoform A, complete
Length = 2452
Score = 556 bits (1433), Expect = e-158
Identities = 309/627 (49%), Positives = 418/627 (66%), Gaps = 24/627 (3%)
Frame = +3
Query: 54 VIGIDLGTTNSCVSVMEGKNPKVVENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNPE 113
VIGIDLGTT SCV V + + +++ N +G R TPS +FT E L+G AK A NPE
Sbjct: 177 VIGIDLGTTYSCVGVYKNGHVEIIANDQGNRITPSW-SFTDS-ERLIGEAAKNLAAVNPE 350
Query: 114 NTISGAKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEAKGQQ-----YSPSQIGAF 168
I KRLIGR+F+D + Q++MK+VPYKIV +G +++ K + +SP +I A
Sbjct: 351 RVIFDVKRLIGRKFEDKEVQRDMKLVPYKIVNK-DGKPYIQEKIKDGETKVFSPEEISAM 527
Query: 169 VLTKMKETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALS 228
+LTKMKETAEA+LGK + AV AYFNDAQRQATKDAG IAGL V RIINEPTAAA++
Sbjct: 528 ILTKMKETAEAFLGKKINDAV----AYFNDAQRQATKDAGVIAGLNVARIINEPTAAAIA 695
Query: 229 YGMNKEG---LIAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLVN 285
YG++K+G I VFDLGGGTFDVSIL I NGVFEV ATNGDT LGGEDFD ++++ +
Sbjct: 696 YGLDKKGGEKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTHLGGEDFDQRIMEYFIK 875
Query: 286 EFKRSESIDLSKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLTR 345
+ D+SKD AL +LR AE+AK LSS Q + + + ++ LTR
Sbjct: 876 LINKKHKKDISKDSRALSKLRREAERAKRALSSQHQVRVEIESLFDGVDFSE----PLTR 1043
Query: 346 SKFEALVNNLIERTKAPCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIF-GKSP 404
++FE L N+L +T P + ++DA + IDE++LVGG TR+PKVQ+++ F GK P
Sbjct: 1044ARFEELNNDLFRKTMGPVKKAMEDAGLQKNQIDEIVLVGGSTRIPKVQQLLKDYFDGKEP 1223
Query: 405 CKGVNPDEAVAMGAALQGGILRGD----VKELLLLDVTPLSLGIETLGGIFTRLINRNTT 460
KGVNPDEAVA GAA+QG IL G+ K++LLLDV PL+LGIET+GG+ T+LI RNT
Sbjct: 1224NKGVNPDEAVAYGAAVQGSILSGEGGEETKDILLLDVAPLTLGIETVGGVMTKLIPRNTV 1403
Query: 461 IPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQIEVTFD 520
IPTKKSQVF+T D QT V I+V +GER + D ++LG+FEL G+PPAPRG PQIEVTF+
Sbjct: 1404IPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGKFELSGIPPAPRGTPQIEVTFE 1583
Query: 521 IDANGIVTVSAKDKSTGKEQQITI-KSSGGLSDDEIQNMVKEAELHAQKDQERKSLIDIR 579
+DANGI+ V A+DK TGK ++ITI G LS +EI+ MV+EAE A++D++ K ID R
Sbjct: 1584VDANGILNVKAEDKGTGKSEKITITNEKGRLSQEEIERMVREAEEFAEEDKKVKERIDAR 1763
Query: 580 NSADTSIYSIEKSLSE---YREKIPAEVAKEIEDAV-------SDLRSAMAGDSADEIKS 629
NS +T +Y+++ +S+ +K+ ++ +++E AV D +S + +++K
Sbjct: 1764NSLETYVYNMKNQVSDKDKLADKLESDEKEKVETAVKEALEWLDDNQSVEKEEYEEKLKE 1943
Query: 630 KLDAANKAVSKIGEHMSGGSSSGPSSD 656
N +S + + G G S +
Sbjct: 1944VEAVCNPIISAVYQRSGGAPGGGASGE 2024
>TC219546 UP|HS70_SOYBN (P26413) Heat shock 70 kDa protein, complete
Length = 2099
Score = 555 bits (1431), Expect = e-158
Identities = 311/629 (49%), Positives = 411/629 (64%), Gaps = 21/629 (3%)
Frame = +1
Query: 55 IGIDLGTTNSCVSVMEGKNPKVVENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNPEN 114
IGIDLGTT SCV V + +++ N +G RTTPS VAFT E L+G AK Q NP+N
Sbjct: 25 IGIDLGTTYSCVGVWQNDRVEIIPNDQGNRTTPSYVAFTDT-ERLIGDAAKNQVAMNPQN 201
Query: 115 TISGAKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAW--VEAKGQQ--YSPSQIGAFVL 170
T+ AKRLIGRRF D Q +MK+ P+K+ +P V KG++ +S +I + VL
Sbjct: 202 TVFDAKRLIGRRFSDSSVQNDMKLWPFKVGGSPCDKPMIVVNYKGEEKKFSAEEISSMVL 381
Query: 171 TKMKETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALSYG 230
KM+E AEA+LG V AV+TVPAYFND+QRQATKDAG I+GL VLRIINEPTAAA++YG
Sbjct: 382 VKMREVAEAFLGHAVKNAVVTVPAYFNDSQRQATKDAGAISGLNVLRIINEPTAAAIAYG 561
Query: 231 MNK------EGLIAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLV 284
++K E + +FDLGGGTFDVSIL I G+FEVKAT GDT LGGEDFDN +++ V
Sbjct: 562 LDKKASRKGEQNVLIFDLGGGTFDVSILTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFV 741
Query: 285 NEFKRSESIDLSKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLT 344
+EFKR D+S + AL+RLR A E+AK LSST+QT I + + T+T
Sbjct: 742 SEFKRKNKKDISGNARALRRLRTACERAKRTLSSTAQTTIEIDSLYEGID----FYATIT 909
Query: 345 RSKFEALVNNLIERTKAPCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIF-GKS 403
R++FE + +L + P + CL+DA I + EV+LVGG TR+PKV +++ F GK
Sbjct: 910 RARFEEMNMDLFRKCMEPVEKCLRDAKIDKSQVHEVVLVGGSTRIPKVHQLLQDFFNGKE 1089
Query: 404 PCKGVNPDEAVAMGAALQGGILRGD----VKELLLLDVTPLSLGIETLGGIFTRLINRNT 459
CK +NPDEAVA GAA+Q IL G V++LLLLDVTPLSLG+ET GG+ T LI RNT
Sbjct: 1090LCKSINPDEAVAYGAAVQAAILSGQGDEKVQDLLLLDVTPLSLGLETAGGVMTVLIPRNT 1269
Query: 460 TIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQIEVTF 519
TIPTKK Q+FST +DNQ V I+V +GER DN +LG+FEL G+PPAPRG+PQI V F
Sbjct: 1270TIPTKKEQIFSTYSDNQPGVLIQVFEGERARTKDNNLLGKFELTGIPPAPRGVPQINVCF 1449
Query: 520 DIDANGIVTVSAKDKSTGKEQQITI-KSSGGLSDDEIQNMVKEAELHAQKDQERKSLIDI 578
DIDANGI+ VSA+DK+ G + +ITI G LS +EI+ MVK+AE + +D+E K ++
Sbjct: 1450DIDANGILNVSAEDKTAGVKNKITITNDKGRLSKEEIEKMVKDAERYKAEDEEVKKKVEA 1629
Query: 579 RNSADTSIYSIEKSLSEYR--EKIPAEVAKEIEDAVSDLRSAMAGD---SADEIKSKLDA 633
+NS + Y++ ++ + + K+ + ++IE AV D + G+ DE + K
Sbjct: 1630KNSLENYAYNMRNTIKDEKIGGKLSPDEKQKIEKAVEDAIQWLEGNQMAEVDEFEDKQKE 1809
Query: 634 ANKAVSKIGEHMSGGSSSGPSSDGSQGGD 662
+ I M G ++GP D G D
Sbjct: 1810LEGICNPIIAKMYQG-AAGPGGDVPMGAD 1893
>TC205463 UP|Q39804 (Q39804) BiP isoform B, complete
Length = 2383
Score = 502 bits (1293), Expect = e-142
Identities = 279/513 (54%), Positives = 358/513 (69%), Gaps = 15/513 (2%)
Frame = +3
Query: 54 VIGIDLGTTNSCVSVMEGKNPKVVENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNPE 113
VIGIDL TT SCV V + +++ N +G R TPS VAFT E L+G AK A NP
Sbjct: 198 VIGIDL-TTYSCVGVTDAV--EIIANDQGNRITPSWVAFTDS-ERLIGEAAKIVAAVNPV 365
Query: 114 NTISGAKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEAKGQQ-----YSPSQIGAF 168
TI KRLIGR+F+D + Q++MK+VPYKIV +G +++ K + +SP +I A
Sbjct: 366 RTIFDVKRLIGRKFEDKEVQRDMKLVPYKIVNK-DGKPYIQVKIKDGETKVFSPEEISAM 542
Query: 169 VLTKMKETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALS 228
+LTKMKETAEA+LGK + AV+TVPAYFNDAQRQATKDAG IAGL V RIINEPTAAA++
Sbjct: 543 ILTKMKETAEAFLGKKINDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAAAIA 722
Query: 229 YGMNKEG---LIAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLVN 285
YG++K+G I VFDLGGGTFDVSIL I NGVFEV ATNGDT LGGEDFD ++++ +
Sbjct: 723 YGLDKKGGEKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTHLGGEDFDQRIMEYFIK 902
Query: 286 EFKRSESIDLSKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLTR 345
K+ D+SKD AL +LR AE+AK LSS Q + + + ++ LTR
Sbjct: 903 LIKKKHGKDISKDNRALGKLRREAERAKRALSSQHQVRVEIESLFDGVDFSE----PLTR 1070
Query: 346 SKFEALVNNLIERTKAPCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIF-GKSP 404
++FE L N+L +T P + ++DA + IDE++LVGG TR+PKVQ+++ F GK P
Sbjct: 1071ARFEELNNDLFRKTMGPVKKAMEDAGLQKSQIDEIVLVGGSTRIPKVQQLLKDYFDGKEP 1250
Query: 405 CKGVNPDEAVAMGAALQGGILRGD----VKELLLLDVTPLSLGIETLGGIFTRLINRNTT 460
KGVNPDEAVA GAA+QG IL G+ K++LLLDV PL+LGIET+GG+ T+LI RNT
Sbjct: 1251NKGVNPDEAVAYGAAVQGSILSGEGGEETKDILLLDVAPLTLGIETVGGVMTKLIPRNTV 1430
Query: 461 IPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQIEVTFD 520
IPTKKSQVF+T D QT V I+V +GER + D ++LG+F+L G+PPAPRG PQIEVTF+
Sbjct: 1431IPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGKFDLSGIPPAPRGTPQIEVTFE 1610
Query: 521 IDA-NGIVTVSA-KDKSTGKEQQITIKSSGGLS 551
+DA GI+ V A + K TGK ++IT G S
Sbjct: 1611VDAERGILNVKARRAKGTGKSEKITDYKRKGTS 1709
Score = 60.1 bits (144), Expect = 3e-09
Identities = 41/137 (29%), Positives = 72/137 (51%), Gaps = 10/137 (7%)
Frame = +1
Query: 538 KEQQITIKSSGGLSDDEIQNMVKEAELHAQKDQERKSLIDIRNSADTSIYSIEKSLSE-- 595
K IT G LS +EI+ MV+EAE A++D++ K ID RNS +T +Y+++ +S+
Sbjct: 1675 KRSPIT-NEKGRLSQEEIERMVREAEEFAEEDKKVKERIDARNSLETYVYNMKNQISDKD 1851
Query: 596 -YREKIPAEVAKEIEDAV-------SDLRSAMAGDSADEIKSKLDAANKAVSKIGEHMSG 647
+K+ ++ ++IE AV D +S D +++K N +S + + SG
Sbjct: 1852 KLADKLESDEKEKIETAVKEALEWLDDNQSMEKEDYEEKLKEVEAVCNPIISAVYQR-SG 2028
Query: 648 GSSSGPSSDGSQGGDQA 664
G+ G + G + D +
Sbjct: 2029 GAPGGGGASGEEDEDDS 2079
>TC204155 homologue to UP|Q84QJ3 (Q84QJ3) Heat shock protein 70, partial
(75%)
Length = 1640
Score = 413 bits (1061), Expect = e-115
Identities = 227/478 (47%), Positives = 314/478 (65%), Gaps = 21/478 (4%)
Frame = +2
Query: 205 KDAGRIAGLEVLRIINEPTAAALSYGMNKEGL------IAVFDLGGGTFDVSILEISNGV 258
KDAG I+GL V+RIINEPTAAA++YG++K+ + +FDLGGGTFDVS+L I G+
Sbjct: 2 KDAGVISGLNVMRIINEPTAAAIAYGLDKKATSSGEKNVLIFDLGGGTFDVSLLTIEEGI 181
Query: 259 FEVKATNGDTFLGGEDFDNALLDFLVNEFKRSESIDLSKDKLALQRLREAAEKAKIELSS 318
FEVKAT GDT LGGEDFDN +++ V EFKR D+S + AL+RLR A E+AK LSS
Sbjct: 182 FEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKNKKDISGNARALRRLRTACERAKRTLSS 361
Query: 319 TSQTEINLPFITADASGAKHLNITLTRSKFEALVNNLIERTKAPCQSCLKDANISTKDID 378
T+QT I + + T+TR++FE L +L + P + CL+DA + +
Sbjct: 362 TAQTTIEIDSLYEGID----FYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVH 529
Query: 379 EVLLVGGMTRVPKVQEVVSTIF-GKSPCKGVNPDEAVAMGAALQGGILRGD----VKELL 433
+V+LVGG TR+PKVQ+++ F GK CK +NPDEAVA GAA+Q IL G+ V++LL
Sbjct: 530 DVVLVGGSTRIPKVQQLLQDFFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLL 709
Query: 434 LLDVTPLSLGIETLGGIFTRLINRNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMASD 493
LLDVTPLS G+ET GG+ T LI RNTTIPTKK QVFST +DNQ V I+V +GER D
Sbjct: 710 LLDVTPLSTGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERTRTRD 889
Query: 494 NKMLGEFELVGLPPAPRGMPQIEVTFDIDANGIVTVSAKDKSTGKEQQITI-KSSGGLSD 552
N +LG+FEL G+PPAPRG+PQI V FDIDANGI+ VSA+DK+TG++ +ITI G LS
Sbjct: 890 NNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSK 1069
Query: 553 DEIQNMVKEAELHAQKDQERKSLIDIRNSADTSIYSIEKSLSEYR--EKIPAEVAKEIED 610
+EI+ MV+EAE + +D+E K ++ +N+ + Y++ ++ + + K+ ++ K+IED
Sbjct: 1070EEIEKMVQEAEKYKSEDEEHKKKVEAKNALENYAYNMRNTIKDDKIASKLSSDDKKKIED 1249
Query: 611 AVSDLRSAMAGD---SADEIKSKL----DAANKAVSKIGEHMSGGSSSGPSSDGSQGG 661
A+ + G+ ADE + K+ N ++K+ + G + DG G
Sbjct: 1250AIEQAIQWLDGNQLAEADEFEDKMKELESICNPIIAKMYQGAGGDAGGAMDEDGPAAG 1423
>TC204160 homologue to UP|HS72_LYCES (P27322) Heat shock cognate 70 kDa
protein 2, partial (66%)
Length = 1390
Score = 400 bits (1029), Expect = e-112
Identities = 218/427 (51%), Positives = 285/427 (66%), Gaps = 15/427 (3%)
Frame = +2
Query: 55 IGIDLGTTNSCVSVMEGKNPKVVENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNPEN 114
IGIDLGTT SCV V + +++ N +G RTTPS VAFT E L+G AK Q NP N
Sbjct: 122 IGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDT-ERLIGDAAKNQVAMNPTN 298
Query: 115 TISGAKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWV----EAKGQQYSPSQIGAFVL 170
T+ AKRLIGRRF D Q +MK+ P+K++ P + + + +Q+S +I + VL
Sbjct: 299 TVFDAKRLIGRRFSDASVQGDMKLWPFKVIPGPAEKPMIVVNYKGEEKQFSAEEISSMVL 478
Query: 171 TKMKETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALSYG 230
KMKE AEAYLG T+ AV+TVPAYFND+QRQATKDAG I+GL V+RIINEPTAAA++YG
Sbjct: 479 MKMKEIAEAYLGSTIKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIINEPTAAAIAYG 658
Query: 231 MNK------EGLIAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLV 284
++K E + +FDLGGGTFDVS+L I G+FEVKAT GDT LGGEDFDN +++ V
Sbjct: 659 LDKKATSSGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFV 838
Query: 285 NEFKRSESIDLSKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLT 344
EFKR D+S + AL+RLR A E+AK LSST+QT I + + T+T
Sbjct: 839 QEFKRKNKKDISGNARALRRLRTACERAKRTLSSTAQTTIEIDSLYEGID----FYTTIT 1006
Query: 345 RSKFEALVNNLIERTKAPCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIF-GKS 403
R++FE L +L + P + CL+DA + + +V+LVGG TR+PKVQ+++ F GK
Sbjct: 1007RARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGSTRIPKVQQLLQDFFNGKE 1186
Query: 404 PCKGVNPDEAVAMGAALQGGILRGD----VKELLLLDVTPLSLGIETLGGIFTRLINRNT 459
CK +NPDEAVA GAA+Q IL G+ V++LLLLDVT LS +ET G+ T L+ NT
Sbjct: 1187LCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTTLSTALETARGVMTVLMTSNT 1366
Query: 460 TIPTKKS 466
TI T+ +
Sbjct: 1367TISTENA 1387
>TC221389 homologue to UP|HS7M_PHAVU (Q01899) Heat shock 70 kDa protein,
mitochondrial precursor, partial (36%)
Length = 827
Score = 202 bits (515), Expect(2) = e-104
Identities = 99/110 (90%), Positives = 102/110 (92%)
Frame = +3
Query: 25 LTGNTKPAYVGHNWSSLSRPFSSRAAGNDVIGIDLGTTNSCVSVMEGKNPKVVENSEGAR 84
LTG+TKPAYV HNWSSLSRPFSSR AGNDVIGIDLGTTNSCVSVMEGKNPKV+ENSEGAR
Sbjct: 165 LTGSTKPAYVAHNWSSLSRPFSSRPAGNDVIGIDLGTTNSCVSVMEGKNPKVIENSEGAR 344
Query: 85 TTPSVVAFTQKGELLVGTPAKRQAVTNPENTISGAKRLIGRRFDDPQTQK 134
TTPSVVAF QKGELLVGTP KRQAVTNP NT+ G KRLIGRRFDD QTQK
Sbjct: 345 TTPSVVAFNQKGELLVGTPXKRQAVTNPTNTLFGTKRLIGRRFDDAQTQK 494
Score = 196 bits (497), Expect(2) = e-104
Identities = 99/112 (88%), Positives = 106/112 (94%), Gaps = 1/112 (0%)
Frame = +1
Query: 133 QKEMKMVPYKIVKAPNGDAWVEAKGQQYSPSQIGAFVLTKMKETAEAYLGKTVPKAVITV 192
++EMKMVP+KIVKAPNG+AWVE GQQYSPSQIGAFVLTKMKETAEAYLGK++ KAVITV
Sbjct: 490 KREMKMVPFKIVKAPNGNAWVETNGQQYSPSQIGAFVLTKMKETAEAYLGKSISKAVITV 669
Query: 193 PAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALSYGMN-KEGLIAVFDLG 243
PAYFNDAQRQATKDAGRIAGL+V RIINEPTAAALSYGMN KEGLIA FDLG
Sbjct: 670 PAYFNDAQRQATKDAGRIAGLDVQRIINEPTAAALSYGMNKKEGLIAXFDLG 825
>TC214818 homologue to UP|Q8GSN4 (Q8GSN4) Non-cell-autonomous heat shock
cognate protein 70, partial (68%)
Length = 1554
Score = 366 bits (939), Expect = e-101
Identities = 207/432 (47%), Positives = 281/432 (64%), Gaps = 12/432 (2%)
Frame = +2
Query: 243 GGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLVNEFKRSESIDLSKDKLAL 302
GGGTFDVS+L I G+FEVKAT GDT LGGEDFDN +++ V EFKR D+S + AL
Sbjct: 8 GGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKNKKDISGNPRAL 187
Query: 303 QRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLTRSKFEALVNNLIERTKAP 362
+RLR A E+AK LSST+QT I + + T+TR++FE L +L + P
Sbjct: 188 RRLRTACERAKRTLSSTAQTTIEIDSLYEGID----FYSTITRARFEELNMDLFRKCMEP 355
Query: 363 CQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIF-GKSPCKGVNPDEAVAMGAALQ 421
+ CL+DA + + + +V+LVGG TR+PKVQ+++ F GK CK +NPDEAVA GAA+Q
Sbjct: 356 VEKCLRDAKMDKRTVHDVVLVGGSTRIPKVQQLLQDFFNGKELCKSINPDEAVAYGAAVQ 535
Query: 422 GGILRGD----VKELLLLDVTPLSLGIETLGGIFTRLINRNTTIPTKKSQVFSTAADNQT 477
IL G+ V++LLLLDVTPLSLG+ET GG+ T LI RNTTIPTKK QVFST +DNQ
Sbjct: 536 AAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQP 715
Query: 478 QVGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQIEVTFDIDANGIVTVSAKDKSTG 537
V I+V +GER DN +LG+FEL G+PPAPRG+PQI V FDIDANGI+ VSA+DK+TG
Sbjct: 716 GVLIQVYEGERTRTRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTG 895
Query: 538 KEQQITI-KSSGGLSDDEIQNMVKEAELHAQKDQERKSLIDIRNSADTSIYSIEKSLSEY 596
++ +ITI G LS +EI+ MV+EAE + +D+E K ++ +N+ + Y++ ++ +
Sbjct: 896 QKNKITITNDKGRLSKEEIEKMVQEAEKYKSEDEEHKKKVEAKNALENYSYNMRNTIKDE 1075
Query: 597 R--EKIPAEVAKEIEDAVSD----LRSAMAGDSADEIKSKLDAANKAVSKIGEHMSGGSS 650
+ K+ K+IEDA+ L S G+ ADE + K+ + I M G
Sbjct: 1076KIGGKLDPADKKKIEDAIEQAIQWLDSNQLGE-ADEFEDKMKELESICNPIIAKMYQG-G 1249
Query: 651 SGPSSDGSQGGD 662
+GP G+ D
Sbjct: 1250AGPDVGGAMDDD 1285
>TC226591 homologue to PRF|1909352A.0|445605|1909352A heat shock protein
hsp70. {Pisum sativum;} , partial (45%)
Length = 1338
Score = 321 bits (823), Expect = 6e-88
Identities = 161/310 (51%), Positives = 216/310 (68%), Gaps = 14/310 (4%)
Frame = +3
Query: 367 LKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIFGKSPCKGVNPDEAVAMGAALQGGILR 426
L+DA +S KD+DEV+LVGG TR+P VQE+V + GK P VNPDE VA+GAA+Q G+L
Sbjct: 3 LRDAKLSFKDLDEVILVGGSTRIPAVQELVKKLTGKDPNVTVNPDEVVALGAAVQAGVLA 182
Query: 427 GDVKELLLLDVTPLSLGIETLGGIFTRLINRNTTIPTKKSQVFSTAADNQTQVGIKVLQG 486
GDV +++LLDVTPLSLG+ETLGG+ T++I RNTT+PT KS+VFSTAAD QT V I VLQG
Sbjct: 183 GDVSDIVLLDVTPLSLGLETLGGVMTKIIPRNTTLPTSKSEVFSTAADGQTSVEINVLQG 362
Query: 487 EREMASDNKMLGEFELVGLPPAPRGMPQIEVTFDIDANGIVTVSAKDKSTGKEQQITIKS 546
ERE DNK LG F L G+PPAPRG+PQIEV FDIDANGI++V+A DK TGK+Q ITI
Sbjct: 363 EREFVRDNKSLGSFRLDGIPPAPRGVPQIEVKFDIDANGILSVTAIDKGTGKKQDITITG 542
Query: 547 SGGLSDDEIQNMVKEAELHAQKDQERKSLIDIRNSADTSIYSIEKSLSEYREKIPAEVAK 606
+ L DE++ MV EAE +++D+E++ ID +N AD+ +Y EK L E +K+P V +
Sbjct: 543 ASTLPSDEVERMVNEAEKFSKEDKEKRDAIDTKNQADSVVYQTEKQLKELGDKVPGPVKE 722
Query: 607 EIEDAVSDLRSAMAGDSADEIKSKLDAANKAVSKIGEHMSG--------------GSSSG 652
++E + +L+ A++G S IK + A N+ V ++G+ + G+ SG
Sbjct: 723 KVEAKLGELKDAISGGSTQAIKDAMAALNQEVMQLGQSLYNQPGAAGAGGPTPPPGADSG 902
Query: 653 PSSDGSQGGD 662
PS +G D
Sbjct: 903 PSESSGKGPD 932
>TC214819 homologue to UP|Q8GSN2 (Q8GSN2) Cell-autonomous heat shock cognate
protein 70, partial (54%)
Length = 1266
Score = 283 bits (723), Expect = 2e-76
Identities = 153/339 (45%), Positives = 222/339 (65%), Gaps = 15/339 (4%)
Frame = +1
Query: 342 TLTRSKFEALVNNLIERTKAPCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIF- 400
T+TR++FE L +L + P + CL+DA + + +D+V+LVGG TR+PKVQ+++ F
Sbjct: 25 TVTRARFEELNMDLFRKCMEPVEKCLRDAKMDKRSVDDVVLVGGSTRIPKVQQLLQDFFN 204
Query: 401 GKSPCKGVNPDEAVAMGAALQGGILRGD----VKELLLLDVTPLSLGIETLGGIFTRLIN 456
GK CK +NPDEAVA GAA+Q IL G+ V++LLLLDVTPLSLG+ET GG+ T LI
Sbjct: 205 GKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIP 384
Query: 457 RNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQIE 516
RNTTIPTKK QVFST +DNQ V I+V +GER DN +LG+FEL G+PPAPRG+PQI
Sbjct: 385 RNTTIPTKKEQVFSTYSDNQPGVLIQVFEGERARTKDNNLLGKFELSGIPPAPRGVPQIT 564
Query: 517 VTFDIDANGIVTVSAKDKSTGKEQQITI-KSSGGLSDDEIQNMVKEAELHAQKDQERKSL 575
V FDIDANGI+ VSA+DK+TG++ +ITI G LS ++I+ MV+EAE + +D+E K
Sbjct: 565 VCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEDIEKMVQEAEKYKSEDEEHKKK 744
Query: 576 IDIRNSADTSIYSIEKSLSEYR--EKIPAEVAKEIEDAVSDL-------RSAMAGDSADE 626
++ +N+ + Y++ ++ + + EK+ K+IEDA+ + A A + D+
Sbjct: 745 VEAKNALENYAYNMRNTVKDDKIGEKLDPTDKKKIEDAIEQAIQWLDSNQLAEADEFEDK 924
Query: 627 IKSKLDAANKAVSKIGEHMSGGSSSGPSSDGSQGGDQAP 665
+K N ++K+ + +G G + ++ AP
Sbjct: 925 MKELESICNPIIAKMYQGGAGPDMGGAGAGAAEDDYAAP 1041
>TC214820 homologue to UP|Q9M6R1 (Q9M6R1) High molecular weight heat shock
protein, partial (54%)
Length = 1272
Score = 235 bits (600), Expect(2) = 4e-71
Identities = 135/286 (47%), Positives = 184/286 (64%), Gaps = 11/286 (3%)
Frame = +2
Query: 391 KVQEVVSTIF-GKSPCKGVNPDEAVAMGAALQGGILRGD----VKELLLLDVTPLSLGIE 445
K Q++V + GK CK +NPDEAVA GAA+Q IL G+ V++LLLLDVTPLSLG+E
Sbjct: 164 K*QQLVQDFYNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLE 343
Query: 446 TLGGIFTRLINRNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGEFELVGL 505
T GG+ T LI RNTTIPTKK QVFST +DNQ V I+V +GER DN +LG+FEL G+
Sbjct: 344 TAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERARTRDNNLLGKFELSGI 523
Query: 506 PPAPRGMPQIEVTFDIDANGIVTVSAKDKSTGKEQQITI-KSSGGLSDDEIQNMVKEAEL 564
PPAPRG+PQI V FDIDANGI+ VSA+DK+TG++ +ITI G LS DEI+ MV+EAE
Sbjct: 524 PPAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKDEIEKMVQEAEK 703
Query: 565 HAQKDQERKSLIDIRNSADTSIYSIEKSLSEYR--EKIPAEVAKEIEDAVSDLRSAMAGD 622
+ +D+E K +D +N+ + Y++ ++ + + K+ + K+IEDA+ + G+
Sbjct: 704 YKAEDEEHKKKVDAKNALENYAYNMRNTIKDEKIASKLSDDDKKKIEDAIESAIQWLDGN 883
Query: 623 ---SADEIKSKLDAANKAVSKIGEHMSGGSSSGPSSDGSQGGDQAP 665
ADE + K+ + I M G+ + P G D P
Sbjct: 884 QLAEADEFEDKMKELESICNPIIAKMYQGAGA-PDMAGGMDEDVPP 1018
Score = 52.0 bits (123), Expect(2) = 4e-71
Identities = 22/54 (40%), Positives = 35/54 (64%)
Frame = +1
Query: 342 TLTRSKFEALVNNLIERTKAPCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEV 395
T+TR++FE L +L + P + CL+DA + + +V+LVGG TR+PKV +
Sbjct: 16 TITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGSTRIPKVAAI 177
>TC205200 similar to UP|HS7M_PEA (P37900) Heat shock 70 kDa protein,
mitochondrial precursor, partial (21%)
Length = 646
Score = 251 bits (640), Expect = 1e-66
Identities = 128/143 (89%), Positives = 136/143 (94%)
Frame = +3
Query: 531 AKDKSTGKEQQITIKSSGGLSDDEIQNMVKEAELHAQKDQERKSLIDIRNSADTSIYSIE 590
AKDKSTGKEQQITI+SSGGLSDDEI+ MVKEAELHAQKDQERK+LIDIRNSADT+IYSIE
Sbjct: 9 AKDKSTGKEQQITIRSSGGLSDDEIEKMVKEAELHAQKDQERKALIDIRNSADTTIYSIE 188
Query: 591 KSLSEYREKIPAEVAKEIEDAVSDLRSAMAGDSADEIKSKLDAANKAVSKIGEHMSGGSS 650
KSL EYREKIP+EVAKEIEDAVSDLR AM+GD+ DEIKSKLDAANKAVSKIGEHMSGGSS
Sbjct: 189 KSLGEYREKIPSEVAKEIEDAVSDLRQAMSGDNVDEIKSKLDAANKAVSKIGEHMSGGSS 368
Query: 651 SGPSSDGSQGGDQAPEAEYEEVK 673
G S+ GSQGGDQAPEAEYEEVK
Sbjct: 369 GGSSAGGSQGGDQAPEAEYEEVK 437
>TC204156 homologue to UP|Q40151 (Q40151) Hsc70 protein, partial (40%)
Length = 965
Score = 205 bits (521), Expect = 6e-53
Identities = 112/243 (46%), Positives = 159/243 (65%), Gaps = 10/243 (4%)
Frame = +2
Query: 429 VKELLLLDVTPLSLGIETLGGIFTRLINRNTTIPTKKSQVFSTAADNQTQVGIKVLQGER 488
V++LLLLDVTPLSLG+ET GG+ T LI RNTTIPTKK QVFST +DNQ V I+V +GER
Sbjct: 17 VQDLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGER 196
Query: 489 EMASDNKMLGEFELVGLPPAPRGMPQIEVTFDIDANGIVTVSAKDKSTGKEQQITI-KSS 547
DN +LG+FEL G+PPAPRG+PQI V FDIDANGI+ VSA+DK+TG++ +ITI
Sbjct: 197 TRTRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDK 376
Query: 548 GGLSDDEIQNMVKEAELHAQKDQERKSLIDIRNSADTSIYSIEKSLSEYR--EKIPAEVA 605
G LS +EI+ MV+EAE + +D+E K ++ +N+ + Y++ ++ + + K+ A+
Sbjct: 377 GRLSKEEIEKMVQEAEKYKAEDEEHKKKVEAKNTLENYAYNMRNTIKDDKIASKLSADDK 556
Query: 606 KEIEDAVSDLRSAMAGD---SADEIKSKL----DAANKAVSKIGEHMSGGSSSGPSSDGS 658
K+IEDA+ + G+ ADE + K+ N ++K+ + G + DG
Sbjct: 557 KKIEDAIEQAIQWLDGNQLAEADEFEDKMKELESICNPIIAKMYQGAGGDAGGAMDEDGP 736
Query: 659 QGG 661
G
Sbjct: 737 AAG 745
>TC205201 similar to UP|HS7M_PEA (P37900) Heat shock 70 kDa protein,
mitochondrial precursor, partial (18%)
Length = 467
Score = 178 bits (451), Expect = 8e-45
Identities = 87/103 (84%), Positives = 93/103 (89%)
Frame = +1
Query: 25 LTGNTKPAYVGHNWSSLSRPFSSRAAGNDVIGIDLGTTNSCVSVMEGKNPKVVENSEGAR 84
LT TK +YVG+ W+SLSRPFSS+ AGNDVIGIDLGTTNSCVSVMEGKNPKV+ENSEGAR
Sbjct: 157 LTSGTKTSYVGNKWASLSRPFSSKPAGNDVIGIDLGTTNSCVSVMEGKNPKVIENSEGAR 336
Query: 85 TTPSVVAFTQKGELLVGTPAKRQAVTNPENTISGAKRLIGRRF 127
TTPSVVAF QK ELLVGTPAKRQAVTNP NT+ G KRLIGRRF
Sbjct: 337 TTPSVVAFNQKAELLVGTPAKRQAVTNPTNTLFGTKRLIGRRF 465
>TC234487 similar to UP|Q9SAU8 (Q9SAU8) HSP70, partial (23%)
Length = 457
Score = 152 bits (385), Expect = 4e-37
Identities = 83/153 (54%), Positives = 104/153 (67%), Gaps = 4/153 (2%)
Frame = +1
Query: 77 VENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNPENTISGAKRLIGRRFDDPQTQKEM 136
+ N +G TTPS VAFT + + L+G AK QA TNPENT+ AKRLIGR+F DP QK+
Sbjct: 1 IHNDQGNNTTPSCVAFTDQ-QRLIGEAAKNQAATNPENTVFDAKRLIGRKFSDPVIQKDK 177
Query: 137 KMVPYKIVKAPNGDAWVEA--KGQQYS--PSQIGAFVLTKMKETAEAYLGKTVPKAVITV 192
+ P+K+V N + KGQ+ ++ + VL KM+E AEAYL V AV+TV
Sbjct: 178 MLWPFKVVAGINDKPMISLNYKGQEKHLLAEEVSSMVLIKMREIAEAYLETPVENAVVTV 357
Query: 193 PAYFNDAQRQATKDAGRIAGLEVLRIINEPTAA 225
PAYFND+QR+AT DAG IAGL V+RIINEPTAA
Sbjct: 358 PAYFNDSQRKATIDAGAIAGLNVMRIINEPTAA 456
>TC222877 weakly similar to UP|Q94614 (Q94614) Heat shock 70kDa protein
(Fragment), partial (19%)
Length = 809
Score = 151 bits (382), Expect = 8e-37
Identities = 95/243 (39%), Positives = 145/243 (59%), Gaps = 17/243 (6%)
Frame = +1
Query: 417 GAALQGGILRG---DVKELLLLDVTPLSLGIETLGGIFTRLINRNTTIPTKKSQVFSTAA 473
GAA+Q +L +V L+LLD+TPLSLG+ G + + +I RNTTIP ++++ + T
Sbjct: 4 GAAVQAALLSKGIVNVPNLVLLDITPLSLGVSVQGDLMSVVIPRNTTIPVRRTKTYVTTE 183
Query: 474 DNQTQVGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQIEVTFDIDANGIVTVSAKD 533
DNQ+ V I+V +GER ASDN +LG F L G+PPAPRG P E TFDID NGI++VSA++
Sbjct: 184 DNQSAVMIEVYEGERTRASDNNLLGFFTLSGIPPAPRGHPLYE-TFDIDENGILSVSAEE 360
Query: 534 KSTGKEQQITI-KSSGGLSDDEIQNMVKEAELHAQKDQERKSLIDIRNSADTSIYSIEKS 592
+STG + +ITI LS EI+ M++EAE + +D++ N D +Y I+ +
Sbjct: 361 ESTGNKNEITITNEKERLSTKEIKRMIQEAEYYKAEDKKFLRKAKAMNDLDYYVYKIKNA 540
Query: 593 L----------SEYREKIPAEVAKE---IEDAVSDLRSAMAGDSADEIKSKLDAANKAVS 639
L S+ +E + + +A+ +ED + D+ E++S ++ KA+
Sbjct: 541 LKKKDISSKLCSKEKENVSSAIARATDLLEDNNQQDDIVVFEDNLKELESIIERM-KAMG 717
Query: 640 KIG 642
KIG
Sbjct: 718 KIG 726
>BQ296289 homologue to PIR|T45517|T45 heat shock protein 70 cytosolic
[imported] - spinach, partial (21%)
Length = 421
Score = 149 bits (376), Expect = 4e-36
Identities = 77/130 (59%), Positives = 97/130 (74%), Gaps = 6/130 (4%)
Frame = +1
Query: 203 ATKDAGRIAGLEVLRIINEPTAAALSYGMNKEGL------IAVFDLGGGTFDVSILEISN 256
ATKDAG IAGL V+RIINEPTAAA++YG++K+ + +FDLGGGTFDVS+L I
Sbjct: 1 ATKDAGVIAGLNVMRIINEPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEE 180
Query: 257 GVFEVKATNGDTFLGGEDFDNALLDFLVNEFKRSESIDLSKDKLALQRLREAAEKAKIEL 316
G+FEVKAT GDT LGGEDFDN +++ V EFKR D++ + AL+RLR A E+AK L
Sbjct: 181 GIFEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKHKKDINGNPRALRRLRTACERAKRTL 360
Query: 317 SSTSQTEINL 326
SST+QT I +
Sbjct: 361 SSTAQTTIEI 390
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.312 0.131 0.355
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,877,281
Number of Sequences: 63676
Number of extensions: 226502
Number of successful extensions: 1019
Number of sequences better than 10.0: 84
Number of HSP's better than 10.0 without gapping: 938
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 940
length of query: 673
length of database: 12,639,632
effective HSP length: 104
effective length of query: 569
effective length of database: 6,017,328
effective search space: 3423859632
effective search space used: 3423859632
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC148396.10


