

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
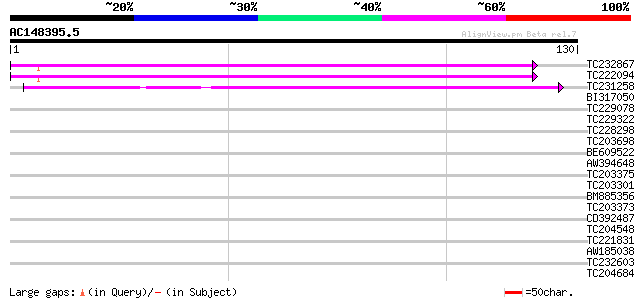
Query= AC148395.5 - phase: 0
(130 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC232867 similar to UP|Q9MA68 (Q9MA68) F2J6.13 protein, partial ... 84 2e-17
TC222094 84 2e-17
TC231258 similar to UP|Q886T6 (Q886T6) Sensory box histidine kin... 54 3e-08
BI317050 29 0.71
TC229078 similar to UP|O82458 (O82458) Rac GTPase activating pro... 28 0.93
TC229322 28 1.6
TC228298 homologue to UP|FRI3_SOYBN (Q948P6) Ferritin 3, chlorop... 28 1.6
TC203698 homologue to UP|Q9XQB0 (Q9XQB0) Carbonic anhydrase, par... 27 2.7
BE609522 similar to GP|16226255|gb| AT3g01500/F4P13_5 {Arabidops... 27 2.7
AW394648 similar to GP|3695059|gb| rac GTPase activating protein... 27 2.7
TC203375 homologue to UP|Q9XQB0 (Q9XQB0) Carbonic anhydrase, par... 27 2.7
TC203301 homologue to UP|Q9XQB0 (Q9XQB0) Carbonic anhydrase, com... 27 2.7
BM885356 27 2.7
TC203373 homologue to UP|Q9XQB0 (Q9XQB0) Carbonic anhydrase, par... 27 2.7
CD392487 27 2.7
TC204548 similar to UP|VT13_ARATH (Q9LVP9) Vesicle transport v-S... 26 4.6
TC221831 UP|Q8LJS1 (Q8LJS1) No apical meristem-like protein, com... 26 6.0
AW185038 25 7.9
TC232603 weakly similar to UP|Q39482 (Q39482) Legumin (Fragment)... 25 7.9
TC204684 similar to UP|RK34_SPIOL (P82244) 50S ribosomal protein... 25 7.9
>TC232867 similar to UP|Q9MA68 (Q9MA68) F2J6.13 protein, partial (3%)
Length = 1144
Score = 83.6 bits (205), Expect = 2e-17
Identities = 41/124 (33%), Positives = 66/124 (53%), Gaps = 3/124 (2%)
Frame = -3
Query: 1 MAEEH---TLKEYATPSTEEPQTVIVYPTIDANNFDIKPALLNIVQQNQFFELPTEDPNL 57
MAEE L++Y++ + + T I P + A P+L+ ++Q N F LP EDP
Sbjct: 929 MAEEPRRVALEDYSSSTVPQFFTSIAQPEVQAQTITYPPSLIQLIQSNFFHGLPNEDPYA 750
Query: 58 HISTFLRLSGTLKSNGVTPETIRLHLFPFLLRDRASAWFHSLPNGSITPWDQLRQAFLAR 117
H++T++ + T++ GV + IRL L F L A W HS S+ WD++ + FL +
Sbjct: 749 HLATYIEICNTIRLAGVPADAIRLSLLSFSLSGEAKRWLHSFKGNSLKSWDEVVEKFLKK 570
Query: 118 SSPK 121
P+
Sbjct: 569 YFPE 558
>TC222094
Length = 984
Score = 83.6 bits (205), Expect = 2e-17
Identities = 42/125 (33%), Positives = 67/125 (53%), Gaps = 4/125 (3%)
Frame = -2
Query: 1 MAEEH----TLKEYATPSTEEPQTVIVYPTIDANNFDIKPALLNIVQQNQFFELPTEDPN 56
MAE H TL++Y++P + T I P + NF +L+ ++Q N F+ LPTEDP
Sbjct: 872 MAENHPQRMTLEDYSSPIIPQYFTSIARPEVQTANFSYPYSLIQLIQGNLFYGLPTEDPY 693
Query: 57 LHISTFLRLSGTLKSNGVTPETIRLHLFPFLLRDRASAWFHSLPNGSITPWDQLRQAFLA 116
H++T++ + T+K GV + I L LF F L A W S ++ W++ + FL
Sbjct: 692 AHLATYIDICNTVKIVGVPEDAIHLDLFCFSLAGEARTWLRSFKGNNLRTWNEXXEKFLK 513
Query: 117 RSSPK 121
+ P+
Sbjct: 512 KYFPE 498
>TC231258 similar to UP|Q886T6 (Q886T6) Sensory box histidine kinase/response
regulator, partial (3%)
Length = 1374
Score = 53.5 bits (127), Expect = 3e-08
Identities = 36/124 (29%), Positives = 55/124 (44%)
Frame = +2
Query: 4 EHTLKEYATPSTEEPQTVIVYPTIDANNFDIKPALLNIVQQNQFFELPTEDPNLHISTFL 63
E TL+E P YP D + +K L++++ + F L EDP+ H+ F
Sbjct: 434 ERTLREMVAPDFTYESLCNQYPDEDVP-YVLKTGLIHLLPK--FHGLVGEDPHKHLKEFH 604
Query: 64 RLSGTLKSNGVTPETIRLHLFPFLLRDRASAWFHSLPNGSITPWDQLRQAFLARSSPKQN 123
+ T+K V + I L FP L A W + L SI WD L++ FL + P
Sbjct: 605 IVCSTMKPPDVQEDHIFLKAFPHSLEGVAKDWLYYLAPRSIFNWDDLKRVFLEKFFPASR 784
Query: 124 CSVV 127
+ +
Sbjct: 785 TTTI 796
>BI317050
Length = 425
Score = 28.9 bits (63), Expect = 0.71
Identities = 17/44 (38%), Positives = 20/44 (44%), Gaps = 1/44 (2%)
Frame = +3
Query: 75 TPETIRLHLFPFLLRDRASAWFHSL-PNGSITPWDQLRQAFLAR 117
TPE RL + F L A +WF L N +T W QA R
Sbjct: 90 TPEDERLQVTSFYLDGEALSWFQWLYRNDQLTSWSSFLQALEMR 221
>TC229078 similar to UP|O82458 (O82458) Rac GTPase activating protein 1,
partial (70%)
Length = 1579
Score = 28.5 bits (62), Expect = 0.93
Identities = 13/37 (35%), Positives = 20/37 (53%)
Frame = +1
Query: 71 SNGVTPETIRLHLFPFLLRDRASAWFHSLPNGSITPW 107
+ GV P+ I +H L++ AWF LP G + P+
Sbjct: 250 NRGVVPDGIDVHCLAGLIK----AWFRELPTGVLDPF 348
>TC229322
Length = 1133
Score = 27.7 bits (60), Expect = 1.6
Identities = 17/60 (28%), Positives = 27/60 (44%), Gaps = 1/60 (1%)
Frame = -3
Query: 23 VYPTIDANNFDIKPALLNIVQQNQF-FELPTEDPNLHISTFLRLSGTLKSNGVTPETIRL 81
++P A D P + + + ELP E N H++T L L + N + E I+L
Sbjct: 780 IFPEASALISDAAPKVGKVCPDHSIKTELPLET*NTHVTTLLHLVAAVIENSLAVEYIQL 601
>TC228298 homologue to UP|FRI3_SOYBN (Q948P6) Ferritin 3, chloroplast
precursor (SFerH-3), partial (94%)
Length = 1250
Score = 27.7 bits (60), Expect = 1.6
Identities = 12/31 (38%), Positives = 15/31 (47%)
Frame = +1
Query: 75 TPETIRLHLFPFLLRDRASAWFHSLPNGSIT 105
TP + P RA+ WFH+LP IT
Sbjct: 412 TPTATIYYSLPLFATLRANTWFHALPRTPIT 504
>TC203698 homologue to UP|Q9XQB0 (Q9XQB0) Carbonic anhydrase, partial (21%)
Length = 1348
Score = 26.9 bits (58), Expect = 2.7
Identities = 17/45 (37%), Positives = 22/45 (48%), Gaps = 5/45 (11%)
Frame = -1
Query: 1 MAEEHTLKEYATP-----STEEPQTVIVYPTIDANNFDIKPALLN 40
M + HT KE A P S+++P T YP N KP+L N
Sbjct: 1186 MVDVHTEKEDARPN*RPQSSKDPLTKS*YPPFSDNVLFTKPSLTN 1052
>BE609522 similar to GP|16226255|gb| AT3g01500/F4P13_5 {Arabidopsis
thaliana}, partial (13%)
Length = 359
Score = 26.9 bits (58), Expect = 2.7
Identities = 17/45 (37%), Positives = 22/45 (48%), Gaps = 5/45 (11%)
Frame = -1
Query: 1 MAEEHTLKEYATP-----STEEPQTVIVYPTIDANNFDIKPALLN 40
M + HT KE A P S+++P T YP N KP+L N
Sbjct: 281 MVDVHTEKEDARPN*RPQSSKDPLTKS*YPPFSDNVLFTKPSLTN 147
>AW394648 similar to GP|3695059|gb| rac GTPase activating protein 1 {Lotus
japonicus}, partial (24%)
Length = 361
Score = 26.9 bits (58), Expect = 2.7
Identities = 12/36 (33%), Positives = 19/36 (52%)
Frame = +2
Query: 71 SNGVTPETIRLHLFPFLLRDRASAWFHSLPNGSITP 106
+ G+ P+ I +H L++ AWF LPN + P
Sbjct: 242 NRGIVPDGIDVHCLAGLIK----AWFRELPNWVLDP 337
>TC203375 homologue to UP|Q9XQB0 (Q9XQB0) Carbonic anhydrase, partial (57%)
Length = 1427
Score = 26.9 bits (58), Expect = 2.7
Identities = 17/45 (37%), Positives = 22/45 (48%), Gaps = 5/45 (11%)
Frame = -2
Query: 1 MAEEHTLKEYATP-----STEEPQTVIVYPTIDANNFDIKPALLN 40
M + HT KE A P S+++P T YP N KP+L N
Sbjct: 925 MVDVHTEKEDARPN*RPQSSKDPLTKS*YPPFSDNVLFTKPSLTN 791
>TC203301 homologue to UP|Q9XQB0 (Q9XQB0) Carbonic anhydrase, complete
Length = 1413
Score = 26.9 bits (58), Expect = 2.7
Identities = 17/45 (37%), Positives = 22/45 (48%), Gaps = 5/45 (11%)
Frame = -2
Query: 1 MAEEHTLKEYATP-----STEEPQTVIVYPTIDANNFDIKPALLN 40
M + HT KE A P S+++P T YP N KP+L N
Sbjct: 1049 MVDVHTEKEDARPN*RPQSSKDPLTKS*YPPFSDNVLFTKPSLTN 915
>BM885356
Length = 420
Score = 26.9 bits (58), Expect = 2.7
Identities = 14/44 (31%), Positives = 22/44 (49%), Gaps = 1/44 (2%)
Frame = -3
Query: 75 TPETIRLHLFPFLLRDRASAWFHSL-PNGSITPWDQLRQAFLAR 117
TP+ RL + F + +A AWF + NG ++ W A +R
Sbjct: 166 TPDHERLTIASFYMEGQALAWFQWMHRNGQLSSWPAFLHALHSR 35
>TC203373 homologue to UP|Q9XQB0 (Q9XQB0) Carbonic anhydrase, partial (38%)
Length = 717
Score = 26.9 bits (58), Expect = 2.7
Identities = 17/45 (37%), Positives = 22/45 (48%), Gaps = 5/45 (11%)
Frame = -3
Query: 1 MAEEHTLKEYATP-----STEEPQTVIVYPTIDANNFDIKPALLN 40
M + HT KE A P S+++P T YP N KP+L N
Sbjct: 391 MVDVHTEKEDARPN*RPQSSKDPLTKS*YPPFSDNVLFTKPSLTN 257
>CD392487
Length = 384
Score = 26.9 bits (58), Expect = 2.7
Identities = 17/45 (37%), Positives = 22/45 (48%), Gaps = 5/45 (11%)
Frame = +3
Query: 1 MAEEHTLKEYATP-----STEEPQTVIVYPTIDANNFDIKPALLN 40
M + HT KE A P S+++P T YP N KP+L N
Sbjct: 54 MVDVHTEKEDARPN*RPQSSKDPLTKS*YPPFSDNVLFTKPSLTN 188
>TC204548 similar to UP|VT13_ARATH (Q9LVP9) Vesicle transport v-SNARE 13
(AtVTI13) (Vesicle transport v-SNARE protein VTI13)
(Vesicle soluble NSF attachment protein receptor 13),
partial (61%)
Length = 657
Score = 26.2 bits (56), Expect = 4.6
Identities = 14/29 (48%), Positives = 15/29 (51%)
Frame = -1
Query: 82 HLFPFLLRDRASAWFHSLPNGSITPWDQL 110
HLF F+L D F LP S TPW L
Sbjct: 342 HLFLFILLDMLVKIFLLLPMLSSTPWSVL 256
>TC221831 UP|Q8LJS1 (Q8LJS1) No apical meristem-like protein, complete
Length = 1216
Score = 25.8 bits (55), Expect = 6.0
Identities = 16/57 (28%), Positives = 22/57 (38%), Gaps = 7/57 (12%)
Frame = -2
Query: 55 PNLHISTFLRLSG-------TLKSNGVTPETIRLHLFPFLLRDRASAWFHSLPNGSI 104
PN IS F + T+ +G P + FP+ R WF L NG +
Sbjct: 1104 PNEEISAFTSVERLVFCETLTISLSGSKPLLLMFDPFPWFSRKALR*WFEELGNGKV 934
>AW185038
Length = 454
Score = 25.4 bits (54), Expect = 7.9
Identities = 11/22 (50%), Positives = 15/22 (68%)
Frame = -2
Query: 75 TPETIRLHLFPFLLRDRASAWF 96
+P + LH+FPFLL AS+ F
Sbjct: 261 SPCVLLLHVFPFLLLQEASSAF 196
>TC232603 weakly similar to UP|Q39482 (Q39482) Legumin (Fragment), partial
(4%)
Length = 1123
Score = 25.4 bits (54), Expect = 7.9
Identities = 12/21 (57%), Positives = 13/21 (61%)
Frame = -2
Query: 68 TLKSNGVTPETIRLHLFPFLL 88
T K + P IRLH FPFLL
Sbjct: 528 TTKLHHFHPSRIRLHPFPFLL 466
>TC204684 similar to UP|RK34_SPIOL (P82244) 50S ribosomal protein L34,
chloroplast precursor, partial (68%)
Length = 864
Score = 25.4 bits (54), Expect = 7.9
Identities = 17/44 (38%), Positives = 20/44 (44%)
Frame = +2
Query: 83 LFPFLLRDRASAWFHSLPNGSITPWDQLRQAFLARSSPKQNCSV 126
LFPFL R R W I WDQ R+ F +Q CS+
Sbjct: 218 LFPFL-RFRLVTWI----GFDI*CWDQKRKRFWPSCEGRQGCSL 334
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.134 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,758,571
Number of Sequences: 63676
Number of extensions: 73744
Number of successful extensions: 367
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 367
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 367
length of query: 130
length of database: 12,639,632
effective HSP length: 86
effective length of query: 44
effective length of database: 7,163,496
effective search space: 315193824
effective search space used: 315193824
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 53 (25.0 bits)
Medicago: description of AC148395.5


