

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148395.10 - phase: 0
(247 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
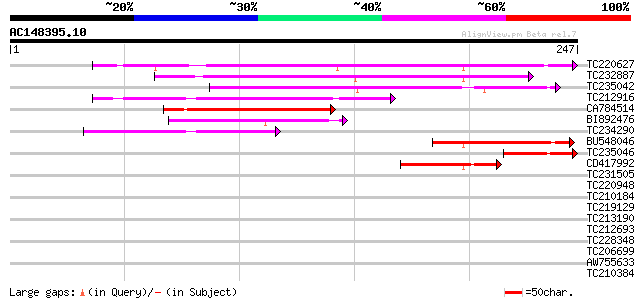
Score E
Sequences producing significant alignments: (bits) Value
TC220627 similar to UP|Q9LI79 (Q9LI79) Gb|AAB84348.1, partial (48%) 152 2e-37
TC232887 129 2e-30
TC235042 120 7e-28
TC212916 similar to UP|Q9FKQ4 (Q9FKQ4) Gb|AAD22354.1 (At5g65340)... 90 1e-18
CA784514 69 3e-12
BI892476 56 2e-08
TC234290 weakly similar to UP|Q9FL47 (Q9FL47) Gb|AAD20416.1, par... 55 3e-08
BU548046 44 7e-05
TC235046 43 2e-04
CD417992 40 8e-04
TC231505 similar to UP|Q9FNY7 (Q9FNY7) 8-oxoguanine DNA glycosyl... 34 0.070
TC220948 similar to UP|Q8IT93 (Q8IT93) Y-box protein Ct-p50, par... 32 0.27
TC210184 weakly similar to GB|AAL57656.1|18086395|AY065011 AT5g1... 30 0.78
TC219129 weakly similar to UP|O23792 (O23792) CND41, chloroplast... 30 1.0
TC213190 homologue to UP|Q7Y0Y9 (Q7Y0Y9) Dehydration responsive ... 30 1.0
TC212693 similar to UP|Q8LFZ9 (Q8LFZ9) Sucrase-like protein, par... 30 1.3
TC228348 weakly similar to UP|EXL2_HUMAN (Q9UBQ6) Exostosin-like... 29 1.7
TC206699 UP|Q7XA53 (Q7XA53) Sucrose transporter, complete 29 2.3
AW755633 weakly similar to GP|15021425|gb| ORF33 {shrimp white s... 29 2.3
TC210384 weakly similar to UP|Q8VZF9 (Q8VZF9) AT5g12890/T24H18_6... 29 2.3
>TC220627 similar to UP|Q9LI79 (Q9LI79) Gb|AAB84348.1, partial (48%)
Length = 1053
Score = 152 bits (383), Expect = 2e-37
Identities = 85/220 (38%), Positives = 130/220 (58%), Gaps = 9/220 (4%)
Frame = +2
Query: 37 NSKSKRSSTNKFFGKFRSMFRSFPII-VPSCKLPTMNGNHRTSETIIHGGTRITGTLFGY 95
N+K K + K KF+S+ F + VP +L ++ + R+ GTLFG+
Sbjct: 341 NAKKKTHFSKK--SKFKSVLTIFTVTRVPRLRLNNISNSFNLD-------MRVMGTLFGH 493
Query: 96 RKARVNLAFQEDSKCHPFLLLELAIPTGKLLQDMGMGLNRIALECE-KHSSNDKTKIVDE 154
R+ V AFQED K P L++LA T L+++M GL RIALECE K ++ K ++++E
Sbjct: 494 RRDHVYFAFQEDPKLGPTFLVKLATRTSTLVREMASGLVRIALECEKKKNATAKVRLLEE 673
Query: 155 PIWTLFCNGKKMGYGVKRDPTDDDLYVIQMLHAVSVAVGVLP-------SDMSDPQDGEL 207
P+W +CNG+K GY +R+ + +++ + +S+ GVLP + + GE+
Sbjct: 674 PLWRTYCNGRKCGYANRRECGPHEWKILKAVEPISMGAGVLPVVCGNEAAGSEEEASGEV 853
Query: 208 SYMRAHFERVIGSKDSETYYMMMPDGNSNGPELSVFFVRV 247
YMRA +ERV+GS+DSE +YMM PD S GPELS++ +RV
Sbjct: 854 MYMRAKYERVVGSRDSEAFYMMNPDA-SGGPELSIYLIRV 970
>TC232887
Length = 555
Score = 129 bits (323), Expect = 2e-30
Identities = 73/187 (39%), Positives = 105/187 (56%), Gaps = 22/187 (11%)
Frame = +2
Query: 64 PSCKLPTMNGNHRTSETIIHGGTRITGTLFGYRKARVNLAFQEDSKCHPFLLLELAIPTG 123
P+CK T+ + ++ G ++TGTLFG+R+ ++ A Q + P LLLELA+ T
Sbjct: 2 PTCKWLTIPSQLSVTPSL---GRKVTGTLFGHRRGHISFAVQLHPRADPVLLLELAMSTS 172
Query: 124 KLLQDMGMGLNRIALECEKHSSNDKT-------------KIVDEPIWTLFCNGKKMGYGV 170
L+++M GL RIALE +K S++ T K+ EP WT++CNG+ GY V
Sbjct: 173 SLVKEMSSGLVRIALESQKLSASTITRTMRSNSGRQQQCKLFQEPSWTMYCNGRNCGYAV 352
Query: 171 KRDPTDDDLYVIQMLHAVSVAVGVLP---------SDMSDPQDGELSYMRAHFERVIGSK 221
R D D +V+ + +VSV GV+P + +GEL YMRA FERV+GS+
Sbjct: 353 SRTCGDLDWHVLSTIQSVSVGAGVIPLLEDGKAASAAAGGGSEGELMYMRARFERVVGSR 532
Query: 222 DSETYYM 228
DSE YM
Sbjct: 533 DSEAXYM 553
>TC235042
Length = 1174
Score = 120 bits (300), Expect = 7e-28
Identities = 65/160 (40%), Positives = 94/160 (58%), Gaps = 7/160 (4%)
Frame = +1
Query: 88 ITGTLFGYRKARVNLAFQEDSKCHPFLLLELAIPTGKLLQDMGMGLNRIALECEKHSSND 147
+ GTLFGYR+ V+ AFQ+D P L+ELA P L+++M GL RIALEC+K ++
Sbjct: 448 LLGTLFGYRRGHVHFAFQKDPTSQPAFLIELATPISGLVREMASGLVRIALECDKDRDSE 627
Query: 148 KTK---IVDEPIWTLFCNGKKMGYGVKRDPTDDDLYVIQMLHAVSVAVGVLPSDMSDPQD 204
K K ++ E +W +CNGKK G+ +R+ D +++ + +S+ GVLP+ D
Sbjct: 628 KKKTLRLLQESVWRTYCNGKKCGFATRRECGAKDWDILKAVEPISMGAGVLPN-----SD 792
Query: 205 G----ELSYMRAHFERVIGSKDSETYYMMMPDGNSNGPEL 240
G + FER+ GS+DSE +YMM PD N PEL
Sbjct: 793 GARW*KSCT*XQXFERIXGSRDSEAFYMMNPDSN-GAPEL 909
>TC212916 similar to UP|Q9FKQ4 (Q9FKQ4) Gb|AAD22354.1 (At5g65340), partial
(12%)
Length = 446
Score = 89.7 bits (221), Expect = 1e-18
Identities = 50/132 (37%), Positives = 78/132 (58%)
Frame = +1
Query: 37 NSKSKRSSTNKFFGKFRSMFRSFPIIVPSCKLPTMNGNHRTSETIIHGGTRITGTLFGYR 96
N +SK SS G M + FP++ CK+ + G R + + TGT+FGYR
Sbjct: 76 NKRSKLSSG----GGLLKMLKLFPMLSSGCKMVALLGRPRK----MLKDSATTGTIFGYR 231
Query: 97 KARVNLAFQEDSKCHPFLLLELAIPTGKLLQDMGMGLNRIALECEKHSSNDKTKIVDEPI 156
K RV+LA QED++ P L+EL + L ++M + RIALE E + ++K K+++E +
Sbjct: 232 KGRVSLAIQEDTRQMPIFLIELPMMASALNKEMASDIVRIALESE--TKSNKKKLLEEFV 405
Query: 157 WTLFCNGKKMGY 168
W ++CNG+K+GY
Sbjct: 406 WAVYCNGRKVGY 441
>CA784514
Length = 435
Score = 68.6 bits (166), Expect = 3e-12
Identities = 36/75 (48%), Positives = 46/75 (61%)
Frame = +2
Query: 68 LPTMNGNHRTSETIIHGGTRITGTLFGYRKARVNLAFQEDSKCHPFLLLELAIPTGKLLQ 127
L + NHR S G+R+ GTLFGYR+ V+ AFQ D P L+ELA P L++
Sbjct: 209 LTVFSKNHR-SNLPFGLGSRVVGTLFGYRRGHVHFAFQRDPTSQPAFLIELATPISGLVR 385
Query: 128 DMGMGLNRIALECEK 142
+M GL RIALEC+K
Sbjct: 386 EMASGLVRIALECDK 430
>BI892476
Length = 406
Score = 55.8 bits (133), Expect = 2e-08
Identities = 33/79 (41%), Positives = 47/79 (58%), Gaps = 1/79 (1%)
Frame = +3
Query: 70 TMNGNHRTSETIIHGGTRITGTLFGYRKARVNLAFQEDSKC-HPFLLLELAIPTGKLLQD 128
T + S +++ T ITGT+FGYR+ +V+ Q ++ +P LLLELA+PT L ++
Sbjct: 171 TSKYTYSPSSSLMSSTTNITGTIFGYRRGKVSFCIQANANSTNPILLLELAVPTTILAKE 350
Query: 129 MGMGLNRIALECEKHSSND 147
M G RIALE S ND
Sbjct: 351 MRGGTLRIALE----SGND 395
>TC234290 weakly similar to UP|Q9FL47 (Q9FL47) Gb|AAD20416.1, partial (11%)
Length = 454
Score = 55.1 bits (131), Expect = 3e-08
Identities = 31/86 (36%), Positives = 47/86 (54%)
Frame = +1
Query: 33 LQPANSKSKRSSTNKFFGKFRSMFRSFPIIVPSCKLPTMNGNHRTSETIIHGGTRITGTL 92
L+ ++KRS+ G M + FP++ CK+ + G R + + TGT+
Sbjct: 193 LRHTTGENKRSNKLSSGGGLLKMLKLFPMLSSGCKMVALLGRPRK----MLKDSATTGTI 360
Query: 93 FGYRKARVNLAFQEDSKCHPFLLLEL 118
FGYRK RV+LA QED++ P L+EL
Sbjct: 361 FGYRKGRVSLAIQEDTRQMPVFLIEL 438
>BU548046
Length = 195
Score = 43.9 bits (102), Expect = 7e-05
Identities = 24/63 (38%), Positives = 41/63 (64%), Gaps = 1/63 (1%)
Frame = -2
Query: 185 LHAVSVAVGVLP-SDMSDPQDGELSYMRAHFERVIGSKDSETYYMMMPDGNSNGPELSVF 243
+ +V V GV+ ++ + +D +L Y+RA F+RV GS + E++ + P+G+ + ELSVF
Sbjct: 188 MRSVVVGTGVMKCKELVNKEDDQLMYLRASFQRVRGSDNCESFXXIDPEGDID-QELSVF 12
Query: 244 FVR 246
F R
Sbjct: 11 FFR 3
>TC235046
Length = 459
Score = 42.7 bits (99), Expect = 2e-04
Identities = 18/32 (56%), Positives = 25/32 (77%)
Frame = +2
Query: 216 RVIGSKDSETYYMMMPDGNSNGPELSVFFVRV 247
R++GS+DSE +YMM PD N PELS++ +RV
Sbjct: 14 RIVGSRDSEAFYMMNPDSN-GAPELSIYLLRV 106
>CD417992
Length = 138
Score = 40.4 bits (93), Expect = 8e-04
Identities = 22/46 (47%), Positives = 32/46 (68%), Gaps = 2/46 (4%)
Frame = -2
Query: 171 KRDPTDDDLYVIQMLHAVSVAVGVLP--SDMSDPQDGELSYMRAHF 214
++ +DD+L+V+Q L VS+ GVLP SD D DGE++Y+RA F
Sbjct: 137 RKQMSDDELHVMQHLRGVSMGAGVLPTSSDHKD-CDGEMTYIRASF 3
>TC231505 similar to UP|Q9FNY7 (Q9FNY7) 8-oxoguanine DNA glycosylase,
partial (65%)
Length = 806
Score = 33.9 bits (76), Expect = 0.070
Identities = 15/33 (45%), Positives = 18/33 (54%)
Frame = +3
Query: 3 PPHAAQTMTLPTAQRPPPAITSPKVPMQISLQP 35
P ++T T PTA PPP T PK+P S P
Sbjct: 6 PSSTSKTATFPTASTPPPTPTPPKLPCLTS*MP 104
>TC220948 similar to UP|Q8IT93 (Q8IT93) Y-box protein Ct-p50, partial (9%)
Length = 442
Score = 32.0 bits (71), Expect = 0.27
Identities = 21/59 (35%), Positives = 31/59 (51%), Gaps = 2/59 (3%)
Frame = +2
Query: 3 PPHAAQTMTLPTAQRPPPAITSPKVPMQISLQPANS--KSKRSSTNKFFGKFRSMFRSF 59
PP T+ TA + T+PK +IS Q +NS ++RS +++ F KFRS F
Sbjct: 170 PPPPLSTLPPATASPTASSSTAPKP*PRISPQSSNSPKATQRSFSHQIFSKFRSSASRF 346
>TC210184 weakly similar to GB|AAL57656.1|18086395|AY065011
AT5g13210/T31B5_30 {Arabidopsis thaliana;} , partial
(37%)
Length = 825
Score = 30.4 bits (67), Expect = 0.78
Identities = 19/43 (44%), Positives = 20/43 (46%)
Frame = -2
Query: 4 PHAAQTMTLPTAQRPPPAITSPKVPMQISLQPANSKSKRSSTN 46
PH QT T P Q+PP S QP NS S SSTN
Sbjct: 245 PHTTQTPTTPMRQKPPWGPHS--YSQTRHKQPNNSSSSPSSTN 123
>TC219129 weakly similar to UP|O23792 (O23792) CND41, chloroplast nucleoid
DNA binding protein precursor, partial (52%)
Length = 1348
Score = 30.0 bits (66), Expect = 1.0
Identities = 17/46 (36%), Positives = 26/46 (55%)
Frame = +2
Query: 1 MAPPHAAQTMTLPTAQRPPPAITSPKVPMQISLQPANSKSKRSSTN 46
++PP A T P+A PPP +TS +P S PA + S S+++
Sbjct: 896 VSPPPPAPLATSPSALPPPPMLTSNTLPFPQS--PAKTPSTVSTSS 1027
>TC213190 homologue to UP|Q7Y0Y9 (Q7Y0Y9) Dehydration responsive element
binding protein, partial (98%)
Length = 733
Score = 30.0 bits (66), Expect = 1.0
Identities = 12/23 (52%), Positives = 15/23 (65%)
Frame = +1
Query: 3 PPHAAQTMTLPTAQRPPPAITSP 25
PPHA T T++ PP A+TSP
Sbjct: 274 PPHAPTTQPFSTSEVPPHALTSP 342
>TC212693 similar to UP|Q8LFZ9 (Q8LFZ9) Sucrase-like protein, partial (19%)
Length = 471
Score = 29.6 bits (65), Expect = 1.3
Identities = 16/49 (32%), Positives = 26/49 (52%), Gaps = 4/49 (8%)
Frame = +2
Query: 2 APPHAAQTMTLPTAQRPPPAITSPKVPMQISLQPANSKSKR----SSTN 46
APP A++T ++PPP P +P +S P ++++ SSTN
Sbjct: 164 APPEASRTTAFSPPEKPPP----PPIPTSVSPAPISARAPLLAPWSSTN 298
>TC228348 weakly similar to UP|EXL2_HUMAN (Q9UBQ6) Exostosin-like 2
(Glucuronyl-galactosyl-proteoglycan
4-alpha-N-acetylglucosaminyltransferase)
(Alpha-1,4-N-acetylhexosaminyltransferase EXTL2)
(Alpha-GalNAcT EXTL2) (EXT-related protein 2) , partial
(13%)
Length = 1278
Score = 29.3 bits (64), Expect = 1.7
Identities = 15/48 (31%), Positives = 24/48 (49%)
Frame = +3
Query: 2 APPHAAQTMTLPTAQRPPPAITSPKVPMQISLQPANSKSKRSSTNKFF 49
+PP A TL +++PP A + P S P +S + SS++ F
Sbjct: 162 SPPTQATASTLRRSRKPPTAAPTAFAPFSTSAAPNSSSAPPSSSSS*F 305
>TC206699 UP|Q7XA53 (Q7XA53) Sucrose transporter, complete
Length = 2053
Score = 28.9 bits (63), Expect = 2.3
Identities = 18/69 (26%), Positives = 32/69 (46%), Gaps = 4/69 (5%)
Frame = +2
Query: 3 PPHAAQTMTLPTAQRPPPAITSPKVPMQISLQPAN----SKSKRSSTNKFFGKFRSMFRS 58
P H A + +++PPPA+++ ++ S ++ S R ST + F R S
Sbjct: 620 PEHTASSTMSSRSRKPPPAMSTARISKAASSSQSHFSQLSPPPRWST*RKFHSRRRRRLS 799
Query: 59 FPIIVPSCK 67
P I +C+
Sbjct: 800 IPTITGACR 826
>AW755633 weakly similar to GP|15021425|gb| ORF33 {shrimp white spot syndrome
virus} [Shrimp white spot syndrome virus], partial (9%)
Length = 439
Score = 28.9 bits (63), Expect = 2.3
Identities = 12/41 (29%), Positives = 21/41 (50%)
Frame = +3
Query: 1 MAPPHAAQTMTLPTAQRPPPAITSPKVPMQISLQPANSKSK 41
++PP+ + PPP+ T+P P + P+ SKS+
Sbjct: 186 ISPPNLLRASPSSPLLHPPPSTTTPTPPPPLKTSPSFSKSR 308
>TC210384 weakly similar to UP|Q8VZF9 (Q8VZF9) AT5g12890/T24H18_60, partial
(11%)
Length = 860
Score = 28.9 bits (63), Expect = 2.3
Identities = 19/69 (27%), Positives = 29/69 (41%), Gaps = 1/69 (1%)
Frame = +3
Query: 3 PPHAAQTMTLPTAQRPPPAITSPKVPMQISLQPANSKSKRSSTNKFFGKFRSM-FRSFPI 61
PP + + ++ PPA SP ++ P + + SS + K S RS+PI
Sbjct: 531 PPSPRNSASSTSSSAAPPASASPATTPSGTISPTAASTPTSSPYRISPKRVSFTARSYPI 710
Query: 62 IVPSCKLPT 70
P PT
Sbjct: 711 TFPKQMAPT 737
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.135 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,227,406
Number of Sequences: 63676
Number of extensions: 157655
Number of successful extensions: 1293
Number of sequences better than 10.0: 60
Number of HSP's better than 10.0 without gapping: 1201
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1269
length of query: 247
length of database: 12,639,632
effective HSP length: 95
effective length of query: 152
effective length of database: 6,590,412
effective search space: 1001742624
effective search space used: 1001742624
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC148395.10


