

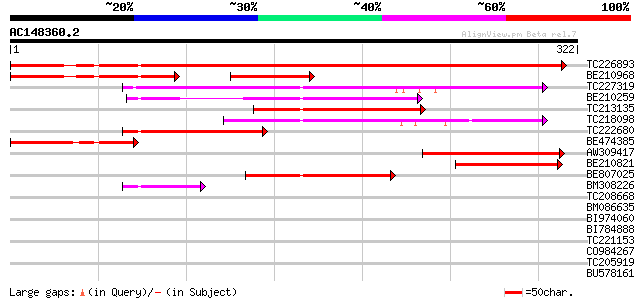
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148360.2 + phase: 0 /pseudo
(322 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC226893 similar to UP|Q9SU94 (Q9SU94) Arginine methyltransferas... 520 e-148
BE210968 120 1e-47
TC227319 similar to UP|Q8W552 (Q8W552) At1g04870/F13M7_12, parti... 150 1e-36
BE210259 similar to GP|9293956|dbj protein arginine N-methyltran... 114 6e-26
TC213135 similar to UP|Q9LJZ9 (Q9LJZ9) Protein arginine N-methyl... 105 4e-23
TC218098 weakly similar to UP|Q8K1Y5 (Q8K1Y5) Carm1-pending prot... 99 2e-21
TC222680 similar to UP|Q9LJZ9 (Q9LJZ9) Protein arginine N-methyl... 94 8e-20
BE474385 89 3e-18
AW309417 87 8e-18
BE210821 85 4e-17
BE807025 similar to GP|10177192|dbj arginine methyltransferase-l... 58 7e-09
BM308226 47 2e-05
TC208668 homologue to UP|Q9NGW8 (Q9NGW8) Developmental protein D... 39 0.003
BM086635 37 0.009
BI974060 37 0.012
BI784888 37 0.016
TC221153 similar to UP|Q9LSF1 (Q9LSF1) Protein kinases-like prot... 36 0.021
CO984267 35 0.046
TC205919 similar to UP|Q94F39 (Q94F39) AT3g29575/MWE13_2, partia... 35 0.060
BU578161 similar to GP|21702253|emb photosystem I subunit O {Ara... 34 0.078
>TC226893 similar to UP|Q9SU94 (Q9SU94) Arginine methyltransferase pam1
(Pam1) , partial (84%)
Length = 1354
Score = 520 bits (1340), Expect = e-148
Identities = 261/317 (82%), Positives = 282/317 (88%), Gaps = 1/317 (0%)
Frame = +2
Query: 1 MGSRKNNNQTNNTNMDNNNNN-NNNHLRFEDADEVIEDVAETSNLDQSMGGCDLDDSNDK 59
MG RKN+N N + + + NNNHLRFE+A V E+SNLDQSM CD+++S+DK
Sbjct: 41 MGQRKNDNNINQCSSSKEDADMNNNHLRFEEA------VDESSNLDQSM--CDIEESDDK 196
Query: 60 TSADYYFDSYSHFGNFHKEMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAGTGILSLFC 119
TSADYYFDSYSHFG H+EMLKDTVRTKTYQNVIYQN+FLFKNKVVLDVGAGTGILSLFC
Sbjct: 197 TSADYYFDSYSHFG-IHEEMLKDTVRTKTYQNVIYQNKFLFKNKVVLDVGAGTGILSLFC 373
Query: 120 AKAGAAHVYAVECSHMADRAKEIVETNGYSKVITVLKGKIEELELPVPKVDIIISEWMGY 179
AKAGA HVYAVECSHMAD AKEIVE NGYS V+TVLKGKIEE+ELPV KVDIIISEWMGY
Sbjct: 374 AKAGAEHVYAVECSHMADMAKEIVEANGYSNVVTVLKGKIEEIELPVAKVDIIISEWMGY 553
Query: 180 FLLFENMLNSVLFARDKWLVDDGVILPDIASLYLTAIEDKDYKEDKIEFWNNVYGFDMSC 239
FLLFENMLNSVL+ARDKWLVD GV+LPD ASL+LTAIED DYKEDKIEFWNNVYGFDMSC
Sbjct: 554 FLLFENMLNSVLYARDKWLVDGGVVLPDKASLHLTAIEDADYKEDKIEFWNNVYGFDMSC 733
Query: 240 IKKQALMEPLVDTVDQNQIATNCQLLKSMDISKMSSGDCSFTAPFKLVAARDDFIHAFVA 299
IKKQA+MEPLVDTVDQNQIATNCQLLK+MDISKM+ GD SF APFKLVA RDD+IHA VA
Sbjct: 734 IKKQAIMEPLVDTVDQNQIATNCQLLKTMDISKMAPGDASFAAPFKLVAERDDYIHALVA 913
Query: 300 YFDVSFTKCHKLMGFST 316
YFDVSFTKCHKLMGFST
Sbjct: 914 YFDVSFTKCHKLMGFST 964
>BE210968
Length = 412
Score = 120 bits (302), Expect(2) = 1e-47
Identities = 64/97 (65%), Positives = 75/97 (76%), Gaps = 1/97 (1%)
Frame = +2
Query: 1 MGSRKNNNQTNNTNMDNNNNN-NNNHLRFEDADEVIEDVAETSNLDQSMGGCDLDDSNDK 59
MG RKN+N N + + + NNNHLRFE+A V E+SNLDQSM CD+++S+DK
Sbjct: 5 MGQRKNDNNINQCSSSKEDADMNNNHLRFEEA------VDESSNLDQSM--CDIEESDDK 160
Query: 60 TSADYYFDSYSHFGNFHKEMLKDTVRTKTYQNVIYQN 96
TSADYYFDSYSHFG H+EMLKDTVRTKTYQNVIYQN
Sbjct: 161TSADYYFDSYSHFG-IHEEMLKDTVRTKTYQNVIYQN 268
Score = 87.4 bits (215), Expect(2) = 1e-47
Identities = 42/48 (87%), Positives = 44/48 (91%)
Frame = +1
Query: 126 HVYAVECSHMADRAKEIVETNGYSKVITVLKGKIEELELPVPKVDIII 173
HVYAVECSHMAD AKEIVE NGYS V+TVLKGKIEE+ELPV KVDIII
Sbjct: 268 HVYAVECSHMADMAKEIVEANGYSNVVTVLKGKIEEIELPVAKVDIII 411
>TC227319 similar to UP|Q8W552 (Q8W552) At1g04870/F13M7_12, partial (87%)
Length = 1540
Score = 150 bits (378), Expect = 1e-36
Identities = 97/264 (36%), Positives = 147/264 (54%), Gaps = 23/264 (8%)
Frame = +2
Query: 65 YFDSYSHFGNFHKEMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAGTGILSLFCAKAGA 124
YF +Y+ F KEML D VR Y + I+QN+ F K VLDVG G+GIL+++ A+AGA
Sbjct: 131 YFCTYA-FLYHQKEMLSDRVRMDAYFDAIFQNKRHFAGKTVLDVGTGSGILAIWSAQAGA 307
Query: 125 AHVYAVECSHMADRAKEIVETNGYSKVITVLKGKIEELELPVPKVDIIISEWMGYFLLFE 184
VYAVE + M++ A+ +V+ N V+ V++G +EE+ LP KVD+IISEWMGYFLL E
Sbjct: 308 RKVYAVEATKMSEHARALVKANNLQDVVEVIEGSMEEITLP-EKVDVIISEWMGYFLLRE 484
Query: 185 NMLNSVLFARDKWLVDDGVILPDIASLYLTAIED--KDYK----EDKIEFWNN------- 231
+M +SV+ ARD+WL GV+ P A +++ I D+K E ++ W++
Sbjct: 485 SMFDSVINARDRWLKPTGVMYPSHARMWMAPIRTGIVDHKLGDYESTMDDWHHFVDETKT 664
Query: 232 VYGFDMSCI--------KKQALMEPLVDTVDQNQIATNCQLLKSMDISKMSSGDC-SFTA 282
YG DMS + +K L L + + +Q+ ++K +D + D +
Sbjct: 665 YYGVDMSTLTRPFSEEQRKYYLQTSLWNNLHPHQVVGTAGIIKEIDCLTATVADIEKVRS 844
Query: 283 PFKLVAARDDF-IHAFVAYFDVSF 305
F + D+ + F +FDV F
Sbjct: 845 DFSMSITVDNTKLCGFGGWFDVHF 916
>BE210259 similar to GP|9293956|dbj protein arginine N-methyltransferase-like
protein {Arabidopsis thaliana}, partial (32%)
Length = 397
Score = 114 bits (285), Expect = 6e-26
Identities = 62/168 (36%), Positives = 94/168 (55%)
Frame = +3
Query: 67 DSYSHFGNFHKEMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAGTGILSLFCAKAGAAH 126
++Y+H G H+EM+KD VRT+TY++ I Q++
Sbjct: 3 EAYAHLG-IHQEMIKDRVRTETYRDAIMQHQ----------------------------- 92
Query: 127 VYAVECSHMADRAKEIVETNGYSKVITVLKGKIEELELPVPKVDIIISEWMGYFLLFENM 186
S +A +A E+V+ N S V+ VL G++E++E+ +VD+IISEWM Y LL+E+M
Sbjct: 93 ------SSIAGKANEVVKANNLSDVVVVLHGRVEDVEID-EEVDVIISEWMSYMLLYESM 251
Query: 187 LNSVLFARDKWLVDDGVILPDIASLYLTAIEDKDYKEDKIEFWNNVYG 234
L SV+ ARD+WL G+ILP A+ Y+ + D ++FW NVYG
Sbjct: 252 LGSVINARDRWLKLGGLILPSSATEYMAPVTHTGRYSDSVDFWRNVYG 395
>TC213135 similar to UP|Q9LJZ9 (Q9LJZ9) Protein arginine
N-methyltransferase-like protein, partial (24%)
Length = 650
Score = 105 bits (261), Expect = 4e-23
Identities = 48/98 (48%), Positives = 69/98 (69%)
Frame = +3
Query: 139 AKEIVETNGYSKVITVLKGKIEELELPVPKVDIIISEWMGYFLLFENMLNSVLFARDKWL 198
A E+V+ N S V+ VL G++E++E+ +VD+IISEWM Y LL+E+ML SV+ ARD+WL
Sbjct: 135 ANEVVKANNLSDVVVVLHGRVEDVEID-EEVDVIISEWMSYMLLYESMLGSVINARDRWL 311
Query: 199 VDDGVILPDIASLYLTAIEDKDYKEDKIEFWNNVYGFD 236
G+ILP A+LY+ + D D+++FW NVYG D
Sbjct: 312 KLGGLILPSSATLYMAPVTHTDRYSDRVDFWRNVYGID 425
>TC218098 weakly similar to UP|Q8K1Y5 (Q8K1Y5) Carm1-pending protein, partial
(6%)
Length = 1369
Score = 99.4 bits (246), Expect = 2e-21
Identities = 66/195 (33%), Positives = 105/195 (53%), Gaps = 11/195 (5%)
Frame = +3
Query: 122 AGAAHVYAVECSHMADRAKEIVETNG-YSKVITVLKGKIEELELPVPKVDIIISEWMGYF 180
AGA HVYAVE S MA+ A++++ N ++ ITV+KGK+E++ELP K DI+ISE MG
Sbjct: 3 AGAKHVYAVEASEMAEYARKLIAGNPTLAQRITVIKGKVEDVELP-EKADILISEPMGTL 179
Query: 181 LLFENMLNSVLFARDKWLVDDGVILPDIASLYLTAIEDKDY---KEDKIEFW--NNVYGF 235
L+ E ML S + ARD++LV G + P + +++ D+ +K FW N YG
Sbjct: 180 LVNERMLESYVIARDRFLVPAGKMFPAVGRIHMAPFTDEYLFIEIANKALFWQQQNYYGV 359
Query: 236 DMSCIKKQALM----EPLVDTVDQNQIATNCQLLKSMDISKMSSGDC-SFTAPFKLVAAR 290
D++ + A +P+VD D + + +D +K+ + P + +A+
Sbjct: 360 DLTPLHGTAFQGYFSQPVVDAFDPRLLIAP-SMFHVIDFTKIKEEELYEIDIPLRFIASV 536
Query: 291 DDFIHAFVAYFDVSF 305
+H +FDV F
Sbjct: 537 GTRVHGLACWFDVLF 581
>TC222680 similar to UP|Q9LJZ9 (Q9LJZ9) Protein arginine
N-methyltransferase-like protein, partial (23%)
Length = 441
Score = 94.0 bits (232), Expect = 8e-20
Identities = 45/82 (54%), Positives = 60/82 (72%)
Frame = +2
Query: 65 YFDSYSHFGNFHKEMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAGTGILSLFCAKAGA 124
YF SY+H G H+EM+KD VRT TY++ I Q++ KVV+DVG GTGILS+FCA+AGA
Sbjct: 194 YFHSYAHLG-IHQEMIKDRVRTDTYRDAIMQHQSFIAGKVVVDVGCGTGILSIFCAQAGA 370
Query: 125 AHVYAVECSHMADRAKEIVETN 146
VYA++ S +A A E+V+ N
Sbjct: 371 KRVYAIDASDIALHANEVVKAN 436
>BE474385
Length = 400
Score = 89.0 bits (219), Expect = 3e-18
Identities = 44/74 (59%), Positives = 57/74 (76%), Gaps = 1/74 (1%)
Frame = +3
Query: 1 MGSRKNNNQTNNTNMDNNNNN-NNNHLRFEDADEVIEDVAETSNLDQSMGGCDLDDSNDK 59
MG RKNN+ N + + + N+NHLRFE+ADE ++ E+SNLDQSM CD+++S+DK
Sbjct: 33 MGRRKNNSNNNQCSSSKEDADMNSNHLRFEEADEAVD---ESSNLDQSM--CDIEESDDK 197
Query: 60 TSADYYFDSYSHFG 73
TSADYYFDSYSHFG
Sbjct: 198 TSADYYFDSYSHFG 239
>AW309417
Length = 445
Score = 87.4 bits (215), Expect = 8e-18
Identities = 46/81 (56%), Positives = 53/81 (64%)
Frame = -1
Query: 235 FDMSCIKKQALMEPLVDTVDQNQIATNCQLLKSMDISKMSSGDCSFTAPFKLVAARDDFI 294
F+MSCIK+ LV V +Q +C LLK MDIS+M GD FT P KLV +I
Sbjct: 445 FNMSCIKQ*RTTPSLVHIVAHHQTVIHC*LLKPMDISQMGPGDDLFTVPLKLVVECAAYI 266
Query: 295 HAFVAYFDVSFTKCHKLMGFS 315
HA VAYFDVSFTKC+KLM FS
Sbjct: 265 HALVAYFDVSFTKCYKLMAFS 203
>BE210821
Length = 282
Score = 85.1 bits (209), Expect = 4e-17
Identities = 40/61 (65%), Positives = 50/61 (81%)
Frame = +3
Query: 254 DQNQIATNCQLLKSMDISKMSSGDCSFTAPFKLVAARDDFIHAFVAYFDVSFTKCHKLMG 313
D+N+IATN QLLK+MD+ +M++GD ++ A KLV RDD+IHA V YFDVS TKCHKLMG
Sbjct: 3 DRNEIATNGQLLKTMDM*EMAAGDAAWGAACKLVGERDDYIHALVGYFDVSCTKCHKLMG 182
Query: 314 F 314
F
Sbjct: 183 F 185
>BE807025 similar to GP|10177192|dbj arginine methyltransferase-like protein
{Arabidopsis thaliana}, partial (14%)
Length = 259
Score = 57.8 bits (138), Expect = 7e-09
Identities = 33/86 (38%), Positives = 56/86 (64%), Gaps = 1/86 (1%)
Frame = +2
Query: 135 MADRAKEIVETNG-YSKVITVLKGKIEELELPVPKVDIIISEWMGYFLLFENMLNSVLFA 193
MA+ A++++ N ++ ITV+KGK+E++ELP K I+I E MG L+ + ML S + A
Sbjct: 5 MAEYARKLIAGNPTLAQRITVIKGKVEDVELP-EKAYILIYEPMGTLLVNKRMLESYVIA 181
Query: 194 RDKWLVDDGVILPDIASLYLTAIEDK 219
D++L +G + PDI +++T D+
Sbjct: 182 IDRFLFPNGKMFPDIGRIHMTPYTDE 259
>BM308226
Length = 428
Score = 46.6 bits (109), Expect = 2e-05
Identities = 24/47 (51%), Positives = 28/47 (59%)
Frame = +1
Query: 65 YFDSYSHFGNFHKEMLKDTVRTKTYQNVIYQNRFLFKNKVVLDVGAG 111
YF SYS FG H+EML D VR Y I +N L + VV+DVG G
Sbjct: 289 YFGSYSSFG-IHREMLSDKVRMDAYGQAILKNPSLLNSAVVMDVGCG 426
>TC208668 homologue to UP|Q9NGW8 (Q9NGW8) Developmental protein DG1037
(Fragment), partial (4%)
Length = 1021
Score = 38.9 bits (89), Expect = 0.003
Identities = 14/25 (56%), Positives = 19/25 (76%)
Frame = -2
Query: 3 SRKNNNQTNNTNMDNNNNNNNNHLR 27
+ NNN NN N +NNNNNNNN+++
Sbjct: 279 NNNNNNNNNNNNNNNNNNNNNNNIK 205
Score = 35.8 bits (81), Expect = 0.027
Identities = 15/37 (40%), Positives = 22/37 (58%)
Frame = -2
Query: 3 SRKNNNQTNNTNMDNNNNNNNNHLRFEDADEVIEDVA 39
+ NNN NN N +NNNNNNN L + ++ + +A
Sbjct: 273 NNNNNNNNNNNNNNNNNNNNNIKLHKKTGSKLNKKIA 163
Score = 30.4 bits (67), Expect = 1.1
Identities = 11/21 (52%), Positives = 16/21 (75%)
Frame = -2
Query: 5 KNNNQTNNTNMDNNNNNNNNH 25
++ + NN N +NNNNNNNN+
Sbjct: 297 QHTSTCNNNNNNNNNNNNNNN 235
Score = 27.7 bits (60), Expect = 7.3
Identities = 10/19 (52%), Positives = 14/19 (73%)
Frame = -2
Query: 7 NNQTNNTNMDNNNNNNNNH 25
+ T+ N +NNNNNNNN+
Sbjct: 300 DQHTSTCNNNNNNNNNNNN 244
>BM086635
Length = 428
Score = 37.4 bits (85), Expect = 0.009
Identities = 14/23 (60%), Positives = 17/23 (73%)
Frame = -2
Query: 3 SRKNNNQTNNTNMDNNNNNNNNH 25
+ NNN NN N +NNNNNNNN+
Sbjct: 229 TNNNNNNNNNNNNNNNNNNNNNN 161
Score = 28.5 bits (62), Expect = 4.3
Identities = 10/19 (52%), Positives = 14/19 (73%)
Frame = -2
Query: 7 NNQTNNTNMDNNNNNNNNH 25
+ + N N +NNNNNNNN+
Sbjct: 241 HTECTNNNNNNNNNNNNNN 185
>BI974060
Length = 420
Score = 37.0 bits (84), Expect = 0.012
Identities = 14/20 (70%), Positives = 16/20 (80%)
Frame = +2
Query: 6 NNNQTNNTNMDNNNNNNNNH 25
NNN NN N +NNNNNNNN+
Sbjct: 269 NNNNNNNNNNNNNNNNNNNN 328
Score = 35.8 bits (81), Expect = 0.027
Identities = 14/21 (66%), Positives = 15/21 (70%)
Frame = +2
Query: 3 SRKNNNQTNNTNMDNNNNNNN 23
S NNN NN N +NNNNNNN
Sbjct: 266 SNNNNNNNNNNNNNNNNNNNN 328
Score = 35.0 bits (79), Expect = 0.046
Identities = 13/20 (65%), Positives = 16/20 (80%)
Frame = +2
Query: 6 NNNQTNNTNMDNNNNNNNNH 25
+NN NN N +NNNNNNNN+
Sbjct: 266 SNNNNNNNNNNNNNNNNNNN 325
Score = 30.0 bits (66), Expect = 1.5
Identities = 11/16 (68%), Positives = 14/16 (86%)
Frame = +2
Query: 10 TNNTNMDNNNNNNNNH 25
T+N N +NNNNNNNN+
Sbjct: 263 TSNNNNNNNNNNNNNN 310
>BI784888
Length = 452
Score = 36.6 bits (83), Expect = 0.016
Identities = 14/19 (73%), Positives = 15/19 (78%)
Frame = +1
Query: 6 NNNQTNNTNMDNNNNNNNN 24
NNN NN N +NNNNNNNN
Sbjct: 373 NNNNNNNNNNNNNNNNNNN 429
Score = 35.0 bits (79), Expect = 0.046
Identities = 14/22 (63%), Positives = 16/22 (72%)
Frame = +1
Query: 4 RKNNNQTNNTNMDNNNNNNNNH 25
R NN NN N +NNNNNNNN+
Sbjct: 364 RGYNNNNNNNNNNNNNNNNNNN 429
Score = 34.7 bits (78), Expect = 0.060
Identities = 14/23 (60%), Positives = 15/23 (64%)
Frame = +1
Query: 1 MGSRKNNNQTNNTNMDNNNNNNN 23
M NNN NN N +NNNNNNN
Sbjct: 361 MRGYNNNNNNNNNNNNNNNNNNN 429
>TC221153 similar to UP|Q9LSF1 (Q9LSF1) Protein kinases-like protein, partial
(38%)
Length = 977
Score = 36.2 bits (82), Expect = 0.021
Identities = 14/29 (48%), Positives = 19/29 (65%)
Frame = +1
Query: 6 NNNQTNNTNMDNNNNNNNNHLRFEDADEV 34
+NN NN N +NNNNNNNN + ++ V
Sbjct: 790 DNNNNNNNNNNNNNNNNNNKVWIDELSHV 876
Score = 31.2 bits (69), Expect = 0.66
Identities = 12/24 (50%), Positives = 15/24 (62%)
Frame = +1
Query: 2 GSRKNNNQTNNTNMDNNNNNNNNH 25
G K +N N +NNNNNNNN+
Sbjct: 763 GGEKTKTLLDNNNNNNNNNNNNNN 834
Score = 30.4 bits (67), Expect = 1.1
Identities = 12/24 (50%), Positives = 15/24 (62%)
Frame = +1
Query: 2 GSRKNNNQTNNTNMDNNNNNNNNH 25
G + NN N +NNNNNNNN+
Sbjct: 766 GEKTKTLLDNNNNNNNNNNNNNNN 837
Score = 28.1 bits (61), Expect = 5.6
Identities = 10/20 (50%), Positives = 14/20 (70%)
Frame = +1
Query: 6 NNNQTNNTNMDNNNNNNNNH 25
+ + T +DNNNNNNNN+
Sbjct: 760 DGGEKTKTLLDNNNNNNNNN 819
Score = 28.1 bits (61), Expect = 5.6
Identities = 10/20 (50%), Positives = 13/20 (65%)
Frame = +1
Query: 1 MGSRKNNNQTNNTNMDNNNN 20
+ + NNN NN N +NNNN
Sbjct: 787 LDNNNNNNNNNNNNNNNNNN 846
>CO984267
Length = 846
Score = 35.0 bits (79), Expect = 0.046
Identities = 25/80 (31%), Positives = 39/80 (48%), Gaps = 5/80 (6%)
Frame = -2
Query: 231 NVYGFDMSCI----KKQALMEPLVDTVDQNQIATNCQLLKSMDISKMSSGDC-SFTAPFK 285
NVYG DMS + K+ A V+T+ + T ++K +D ++ + S TA FK
Sbjct: 845 NVYGIDMSAMVSLAKQCAFEXXXVETITGENVLTWPHVVKYIDSYSVTVQELESVTAKFK 666
Query: 286 LVAARDDFIHAFVAYFDVSF 305
+ +H F +FDV F
Sbjct: 665 FNSMMRAPLHGFAFWFDVEF 606
>TC205919 similar to UP|Q94F39 (Q94F39) AT3g29575/MWE13_2, partial (51%)
Length = 1558
Score = 34.7 bits (78), Expect = 0.060
Identities = 19/40 (47%), Positives = 23/40 (57%)
Frame = +3
Query: 11 NNTNMDNNNNNNNNHLRFEDADEVIEDVAETSNLDQSMGG 50
NN N +NNN+NNNNH +E+ E S LD SM G
Sbjct: 390 NNNNNNNNNSNNNNHHNLPMPVVKLEEELELS-LDLSMNG 506
Score = 28.5 bits (62), Expect = 4.3
Identities = 10/17 (58%), Positives = 13/17 (75%)
Frame = +3
Query: 6 NNNQTNNTNMDNNNNNN 22
NNN NN N +NNN++N
Sbjct: 390 NNNNNNNNNSNNNNHHN 440
>BU578161 similar to GP|21702253|emb photosystem I subunit O {Arabidopsis
thaliana}, partial (43%)
Length = 409
Score = 34.3 bits (77), Expect = 0.078
Identities = 13/18 (72%), Positives = 14/18 (77%)
Frame = -2
Query: 6 NNNQTNNTNMDNNNNNNN 23
NNN NN N +NNNNNNN
Sbjct: 348 NNNNNNNNNNNNNNNNNN 295
Score = 34.3 bits (77), Expect = 0.078
Identities = 13/18 (72%), Positives = 14/18 (77%)
Frame = -2
Query: 7 NNQTNNTNMDNNNNNNNN 24
NN NN N +NNNNNNNN
Sbjct: 348 NNNNNNNNNNNNNNNNNN 295
Score = 33.1 bits (74), Expect = 0.17
Identities = 13/23 (56%), Positives = 17/23 (73%)
Frame = -2
Query: 3 SRKNNNQTNNTNMDNNNNNNNNH 25
S++ N NN N +NNNNNNNN+
Sbjct: 363 SKEPLNNNNNNNNNNNNNNNNNN 295
Score = 30.4 bits (67), Expect = 1.1
Identities = 12/19 (63%), Positives = 15/19 (78%)
Frame = -2
Query: 11 NNTNMDNNNNNNNNHLRFE 29
NN N +NNNNNNNN+ + E
Sbjct: 342 NNNNNNNNNNNNNNNNKKE 286
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.135 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,593,620
Number of Sequences: 63676
Number of extensions: 173430
Number of successful extensions: 3082
Number of sequences better than 10.0: 140
Number of HSP's better than 10.0 without gapping: 1451
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2181
length of query: 322
length of database: 12,639,632
effective HSP length: 97
effective length of query: 225
effective length of database: 6,463,060
effective search space: 1454188500
effective search space used: 1454188500
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC148360.2


