

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148360.13 + phase: 0
(217 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
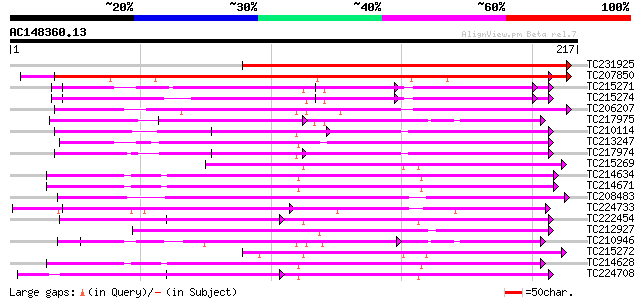
Score E
Sequences producing significant alignments: (bits) Value
TC231925 219 1e-57
TC207850 similar to UP|Q9MA27 (Q9MA27) T5E21.6, partial (63%) 200 5e-52
TC215271 similar to PIR|T01729|T01729 mitochondrial solute carri... 103 5e-23
TC215274 similar to PIR|T01729|T01729 mitochondrial solute carri... 101 2e-22
TC206207 similar to UP|Q8LCH4 (Q8LCH4) Ca-dependent solute carri... 97 7e-21
TC217975 similar to UP|Q9LY28 (Q9LY28) Peroxisomal Ca-dependent ... 90 7e-19
TC210114 similar to UP|Q9FI43 (Q9FI43) Calcium-binding transport... 89 2e-18
TC213247 weakly similar to UP|SFC1_YEAST (P33303) Succinate/fuma... 88 3e-18
TC217974 similar to UP|Q9LY28 (Q9LY28) Peroxisomal Ca-dependent ... 86 1e-17
TC215269 similar to PIR|T01729|T01729 mitochondrial solute carri... 86 2e-17
TC214634 homologue to UP|O49447 (O49447) ADP, ATP carrier-like p... 79 1e-15
TC214671 homologue to UP|O49447 (O49447) ADP, ATP carrier-like p... 79 1e-15
TC208483 similar to GB|AAP21144.1|30102452|BT006336 At5g01500/F7... 79 2e-15
TC224733 similar to GB|AAP12888.1|30017309|BT006239 At5g48970 {A... 77 5e-15
TC222454 similar to UP|Q9LIF7 (Q9LIF7) Mitochondrial carrier pro... 77 6e-15
TC212927 similar to UP|Q9ZNY4 (Q9ZNY4) Mitochondrial energy tran... 77 8e-15
TC210946 similar to UP|Q9FI43 (Q9FI43) Calcium-binding transport... 77 8e-15
TC215272 similar to PIR|T01729|T01729 mitochondrial solute carri... 74 7e-14
TC214628 homologue to UP|O49875 (O49875) Adenine nucleotide tran... 72 2e-13
TC224708 similar to GB|AAP12888.1|30017309|BT006239 At5g48970 {A... 70 6e-13
>TC231925
Length = 751
Score = 219 bits (557), Expect = 1e-57
Identities = 109/126 (86%), Positives = 115/126 (90%)
Frame = +3
Query: 90 LFGIFPYAGLKFYFYEEMKRHVPEDYKKSIMAKLTCGSVAGLLGQTFTYFLEVVRRQMQV 149
L GIFPYAGLKFYFYEEMKRHVPE+Y KSIMAKLTCGSVAGLLGQT TY LEVVRRQMQV
Sbjct: 6 LVGIFPYAGLKFYFYEEMKRHVPEEYNKSIMAKLTCGSVAGLLGQTITYPLEVVRRQMQV 185
Query: 150 QNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYLR 209
Q L S+ AELKGT++S+V IAQKQGWK LFSGLSINYIKVVPS AIGFTVYD+MKSYLR
Sbjct: 186 QKLLPSDNAELKGTLKSVVFIAQKQGWKQLFSGLSINYIKVVPSVAIGFTVYDSMKSYLR 365
Query: 210 VPSRDE 215
VPSRDE
Sbjct: 366 VPSRDE 383
>TC207850 similar to UP|Q9MA27 (Q9MA27) T5E21.6, partial (63%)
Length = 1429
Score = 200 bits (508), Expect = 5e-52
Identities = 102/203 (50%), Positives = 139/203 (68%), Gaps = 5/203 (2%)
Frame = +2
Query: 18 ELLAGGLAGGFAKTVVAPLE--RLKILFQVVSPTKLNVS-GMVNNEQVYRGIRDCLSKTY 74
+LLAG AGG + PL+ R K+ +QV ++ GM + + GI+ L+ Y
Sbjct: 557 DLLAGSAAGGTSVLCTYPLDLARTKLAYQVADTRGGSIKDGMKGVQPAHNGIKGVLTSVY 736
Query: 75 KEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKKSIMAKLTCGSVAGLLGQ 134
KEGG++G+YRG PTL GI PYAGLKFY YE++K HVPE+++KSIM +L+CG++AGL GQ
Sbjct: 737 KEGGVRGLYRGAGPTLTGILPYAGLKFYMYEKLKTHVPEEHQKSIMMRLSCGALAGLFGQ 916
Query: 135 TFTYFLEVVRRQMQVQNL--PASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVP 192
T TY L+VV+RQMQV +L A E+ K T+ + I QGWK LF G+SINYI++VP
Sbjct: 917 TLTYPLDVVKRQMQVGSLQNAAHEDVRYKNTIDGLRTIVCNQGWKQLFHGVSINYIRIVP 1096
Query: 193 SAAIGFTVYDTMKSYLRVPSRDE 215
SAAI FT YD +KS+L +P + +
Sbjct: 1097SAAISFTTYDMVKSWLGIPPQQK 1165
Score = 91.3 bits (225), Expect = 3e-19
Identities = 62/218 (28%), Positives = 105/218 (47%), Gaps = 14/218 (6%)
Frame = +2
Query: 5 SESILDHIPLFAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYR 64
+ES D +P++ KEL+AGG AG +KT VAPLER+KIL+Q +P ++
Sbjct: 230 NESSFDGVPVYVKELIAGGFAGALSKTSVAPLERVKILWQTRTPGFHSL----------- 376
Query: 65 GIRDCLSKTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYK---KSIMA 121
G+ ++K K G G+Y+G ++ I PYA L F YE K + +Y
Sbjct: 377 GVYQSMNKLLKHEGFLGLYKGNGASVIRIVPYAALHFMTYERYKSWILNNYPALGTGPFI 556
Query: 122 KLTCGSVAGLLGQTFTYFLEVVRRQMQVQNLPASEEAELKGTMRS-----------MVLI 170
L GS AG TY L++ R ++ Q + + +K M+ + +
Sbjct: 557 DLLAGSAAGGTSVLCTYPLDLARTKLAYQ-VADTRGGSIKDGMKGVQPAHNGIKGVLTSV 733
Query: 171 AQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYL 208
++ G + L+ G ++P A + F +Y+ +K+++
Sbjct: 734 YKEGGVRGLYRGAGPTLTGILPYAGLKFYMYEKLKTHV 847
Score = 37.0 bits (84), Expect = 0.007
Identities = 30/115 (26%), Positives = 49/115 (42%), Gaps = 3/115 (2%)
Frame = +2
Query: 10 DHIPLFAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDC 69
+H L G LAG F +T+ PL+ +K QV S + + + Y+ D
Sbjct: 854 EHQKSIMMRLSCGALAGLFGQTLTYPLDVVKRQMQVGS-----LQNAAHEDVRYKNTIDG 1018
Query: 70 LSKTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHV---PEDYKKSIMA 121
L G K ++ GV+ I P A + F Y+ +K + P+ +S+ A
Sbjct: 1019LRTIVCNQGWKQLFHGVSINYIRIVPSAAISFTTYDMVKSWLGIPPQQKSQSVSA 1183
>TC215271 similar to PIR|T01729|T01729 mitochondrial solute carrier protein
homolog - Arabidopsis thaliana {Arabidopsis thaliana;} ,
partial (78%)
Length = 1089
Score = 103 bits (258), Expect = 5e-23
Identities = 59/201 (29%), Positives = 107/201 (52%), Gaps = 9/201 (4%)
Frame = +3
Query: 17 KELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKE 76
K L+AGG+AGG ++T VAPLERLKIL QV +P + +G V L ++
Sbjct: 333 KSLVAGGVAGGVSRTAVAPLERLKILLQVQNPHNIKYNGTVQG----------LKYIWRT 482
Query: 77 GGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKKSI---------MAKLTCGS 127
G +G+++G I P + +KF+ YE+ + + YK+ + +L G+
Sbjct: 483 EGFRGLFKGNGTNCARIVPNSAVKFFSYEQASKGILHLYKQQTGNEDAQLTPLLRLGAGA 662
Query: 128 VAGLLGQTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINY 187
AG++ + TY +++VR ++ VQ + + +G ++ + +++G + L+ G +
Sbjct: 663 CAGIIAMSATYPMDMVRGRITVQT--EASPYQYRGMFHALSTVLREEGPRALYKGWLPSV 836
Query: 188 IKVVPSAAIGFTVYDTMKSYL 208
I V+P + F VY+++K YL
Sbjct: 837 IGVIPYVGLNFAVYESLKDYL 899
Score = 75.9 bits (185), Expect = 1e-14
Identities = 44/137 (32%), Positives = 73/137 (53%), Gaps = 8/137 (5%)
Frame = +3
Query: 21 AGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIK 80
AG AG A + P++ ++ ++ V + Q YRG+ LS +E G +
Sbjct: 654 AGACAGIIAMSATYPMDMVR--------GRITVQTEASPYQ-YRGMFHALSTVLREEGPR 806
Query: 81 GIYRGVAPTLFGIFPYAGLKFYFYEEMKRHV--------PEDYKKSIMAKLTCGSVAGLL 132
+Y+G P++ G+ PY GL F YE +K ++ E+ + S+ +L CG+ AG +
Sbjct: 807 ALYKGWLPSVIGVIPYVGLNFAVYESLKDYLIKSNPFDLVENSELSVTTRLACGAAAGTV 986
Query: 133 GQTFTYFLEVVRRQMQV 149
GQT Y L+V+RR+MQ+
Sbjct: 987 GQTVAYPLDVIRRRMQM 1037
Score = 45.4 bits (106), Expect = 2e-05
Identities = 27/85 (31%), Positives = 46/85 (53%)
Frame = +3
Query: 118 SIMAKLTCGSVAGLLGQTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWK 177
SI L G VAG + +T LE ++ +QVQN + GT++ + I + +G++
Sbjct: 324 SICKSLVAGGVAGGVSRTAVAPLERLKILLQVQN---PHNIKYNGTVQGLKYIWRTEGFR 494
Query: 178 TLFSGLSINYIKVVPSAAIGFTVYD 202
LF G N ++VP++A+ F Y+
Sbjct: 495 GLFKGNGTNCARIVPNSAVKFFSYE 569
>TC215274 similar to PIR|T01729|T01729 mitochondrial solute carrier protein
homolog - Arabidopsis thaliana {Arabidopsis thaliana;} ,
partial (77%)
Length = 1034
Score = 101 bits (252), Expect = 2e-22
Identities = 57/201 (28%), Positives = 106/201 (52%), Gaps = 9/201 (4%)
Frame = +2
Query: 17 KELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKE 76
K L+AGG+AGG ++T VAPLERLKIL QV +P + +G + L ++
Sbjct: 272 KSLVAGGVAGGVSRTAVAPLERLKILLQVQNPHSIKYNGTIQG----------LKYIWRT 421
Query: 77 GGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKKSI---------MAKLTCGS 127
G +G+++G I P + +KF+ YE+ + + Y+K + +L G+
Sbjct: 422 EGFRGLFKGNGTNCARIVPNSAVKFFSYEQASKGILHLYRKQTGNEDAQLTPLLRLGAGA 601
Query: 128 VAGLLGQTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINY 187
AG++ + TY +++VR ++ VQ + +G ++ + +++G + L+ G +
Sbjct: 602 CAGIIAMSATYPMDMVRGRITVQT--EKSPYQYRGMFHALSTVLREEGPRALYKGWLPSV 775
Query: 188 IKVVPSAAIGFTVYDTMKSYL 208
I V+P + F VY+++K +L
Sbjct: 776 IGVIPYVGLNFAVYESLKDWL 838
Score = 75.1 bits (183), Expect = 2e-14
Identities = 43/137 (31%), Positives = 70/137 (50%), Gaps = 8/137 (5%)
Frame = +2
Query: 21 AGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIK 80
AG AG A + P++ ++ V + + YRG+ LS +E G +
Sbjct: 593 AGACAGIIAMSATYPMDMVRGRITVQTE---------KSPYQYRGMFHALSTVLREEGPR 745
Query: 81 GIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVP--------EDYKKSIMAKLTCGSVAGLL 132
+Y+G P++ G+ PY GL F YE +K + +D + S+ +L CG+ AG +
Sbjct: 746 ALYKGWLPSVIGVIPYVGLNFAVYESLKDWLVKSNPLGLVQDSELSVTTRLACGAAAGTI 925
Query: 133 GQTFTYFLEVVRRQMQV 149
GQT Y L+V+RR+MQ+
Sbjct: 926 GQTVAYPLDVIRRRMQM 976
Score = 44.3 bits (103), Expect = 4e-05
Identities = 26/85 (30%), Positives = 46/85 (53%)
Frame = +2
Query: 118 SIMAKLTCGSVAGLLGQTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWK 177
+I L G VAG + +T LE ++ +QVQN + GT++ + I + +G++
Sbjct: 263 TICKSLVAGGVAGGVSRTAVAPLERLKILLQVQN---PHSIKYNGTIQGLKYIWRTEGFR 433
Query: 178 TLFSGLSINYIKVVPSAAIGFTVYD 202
LF G N ++VP++A+ F Y+
Sbjct: 434 GLFKGNGTNCARIVPNSAVKFFSYE 508
>TC206207 similar to UP|Q8LCH4 (Q8LCH4) Ca-dependent solute carrier-like
protein, partial (62%)
Length = 1279
Score = 96.7 bits (239), Expect = 7e-21
Identities = 68/209 (32%), Positives = 109/209 (51%), Gaps = 11/209 (5%)
Frame = +2
Query: 18 ELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYR--GIRDCLSKTYK 75
+LLAGG+AG F+KT APL RL ILFQ + GM +N R I + S+
Sbjct: 413 QLLAGGVAGAFSKTCTAPLARLTILFQ--------IQGMHSNVAALRKVSIWNEASRIIH 568
Query: 76 EGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMK---RHVP--EDYKKSIMAKLTC----G 126
E G + ++G T+ PY+ + FY YE K + VP + ++ ++ A L G
Sbjct: 569 EEGFRAFWKGNLVTIAHRLPYSSVNFYSYEHYKKLLKMVPRLQSHRDNVSADLCVHFVGG 748
Query: 127 SVAGLLGQTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSIN 186
+AG+ T TY L++VR ++ Q + +G ++ I++++G L+ GL
Sbjct: 749 GMAGITAATSTYPLDLVRTRLAAQ----TNFTYYRGIWHALHTISKEEGIFGLYKGLGTT 916
Query: 187 YIKVVPSAAIGFTVYDTMKSYLRVPSRDE 215
+ V PS AI F+VY+T++SY + D+
Sbjct: 917 LLTVGPSIAISFSVYETLRSYWQSNRSDD 1003
>TC217975 similar to UP|Q9LY28 (Q9LY28) Peroxisomal Ca-dependent solute
carrier-like protein, partial (44%)
Length = 1187
Score = 90.1 bits (222), Expect = 7e-19
Identities = 57/194 (29%), Positives = 101/194 (51%), Gaps = 4/194 (2%)
Frame = +1
Query: 16 AKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYK 75
+K LAGG+AGG ++T APL+RLK++ QV S I ++K +K
Sbjct: 25 SKYFLAGGIAGGISRTATAPLDRLKVVLQVQSEP--------------ASIMPAVTKIWK 162
Query: 76 EGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDY--KKSI--MAKLTCGSVAGL 131
+ G+ G +RG + + P + +KFY +E +K+ + E + K I +L G AG
Sbjct: 163 QDGLLGFFRGNGLNVVKVSPESAIKFYAFEMLKKVIGEAHGNKSDIGTAGRLVAGGTAGA 342
Query: 132 LGQTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVV 191
+ Q Y +++++ ++Q + +L GT+ + + ++G + + GL + + ++
Sbjct: 343 IAQAAIYPMDLIKTRLQTCPSEGGKVPKL-GTLTMNIWV--QEGPRAFYRGLVPSLLGMI 513
Query: 192 PSAAIGFTVYDTMK 205
P AAI T YDTMK
Sbjct: 514 PYAAIDLTAYDTMK 555
Score = 42.0 bits (97), Expect = 2e-04
Identities = 17/57 (29%), Positives = 30/57 (51%)
Frame = +3
Query: 58 NNEQVYRGIRDCLSKTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPED 114
N Y+G+ D +T++ G G Y+G+ P L + P A + + YE +K+ + D
Sbjct: 696 NTSDAYKGMFDAFRRTFQLEGFIGFYKGLFPNLLKVVPAASITYVVYESLKKTLDLD 866
Score = 38.5 bits (88), Expect = 0.002
Identities = 20/61 (32%), Positives = 34/61 (54%)
Frame = +3
Query: 148 QVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSY 207
Q+Q P++ KG + Q +G+ + GL N +KVVP+A+I + VY+++K
Sbjct: 675 QLQAQPSNTSDAYKGMFDAFRRTFQLEGFIGFYKGLFPNLLKVVPAASITYVVYESLKKT 854
Query: 208 L 208
L
Sbjct: 855 L 857
>TC210114 similar to UP|Q9FI43 (Q9FI43) Calcium-binding transporter-like
protein, partial (28%)
Length = 753
Score = 89.0 bits (219), Expect = 2e-18
Identities = 48/135 (35%), Positives = 78/135 (57%), Gaps = 4/135 (2%)
Frame = +2
Query: 78 GIKGIYRGVAPTLFGIFPYAGLKFYFYEEMK----RHVPEDYKKSIMAKLTCGSVAGLLG 133
G + YRG+ P+L G+ PYAG+ Y+ +K R++ D + +L CG+V+G LG
Sbjct: 38 GPRAFYRGLVPSLLGMIPYAGIDLTAYDTLKDLSKRYILYDSDPGPLVQLGCGTVSGALG 217
Query: 134 QTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPS 193
T Y L+V+R ++Q Q PA+ + KG + +G++ + GL N +KVVP+
Sbjct: 218 ATCVYPLQVIRTRLQAQ--PANSTSAYKGMSDVFWKTLKDEGFRGFYKGLIPNLLKVVPA 391
Query: 194 AAIGFTVYDTMKSYL 208
A+I + VY++MK L
Sbjct: 392 ASITYMVYESMKKSL 436
Score = 53.9 bits (128), Expect = 5e-08
Identities = 31/106 (29%), Positives = 51/106 (47%)
Frame = +2
Query: 18 ELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEG 77
+L G ++G T V PL+ ++ Q N+ Y+G+ D KT K+
Sbjct: 182 QLGCGTVSGALGATCVYPLQVIRTRLQAQP---------ANSTSAYKGMSDVFWKTLKDE 334
Query: 78 GIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKKSIMAKL 123
G +G Y+G+ P L + P A + + YE MK+ + D S++ KL
Sbjct: 335 GFRGFYKGLIPNLLKVVPAASITYMVYESMKKSL--DLD*SVIVKL 466
>TC213247 weakly similar to UP|SFC1_YEAST (P33303) Succinate/fumarate
mitochondrial transporter (Regulator of acetyl-CoA
synthetase activity), partial (11%)
Length = 829
Score = 87.8 bits (216), Expect = 3e-18
Identities = 57/193 (29%), Positives = 94/193 (48%), Gaps = 4/193 (2%)
Frame = +3
Query: 20 LAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGI 79
+ GGL+G + + PL+ ++ T+L + YRGI S ++ G
Sbjct: 84 VGGGLSGITSASATYPLDLVR--------TRLAAQ---RSTMYYRGISHAFSTICRDEGF 230
Query: 80 KGIYRGVAPTLFGIFPYAGLKFYFYEEMKR----HVPEDYKKSIMAKLTCGSVAGLLGQT 135
G+Y+G+ TL G+ P + F YE ++ P+D K + L CGS++G+ T
Sbjct: 231 LGLYKGLGATLLGVGPSIAISFAVYEWLRSVWQSQRPDDSKAVV--GLACGSLSGIASST 404
Query: 136 FTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAA 195
T+ L++VRR+MQ++ + G + I Q +G + L+ G+ Y KVVP
Sbjct: 405 ATFPLDLVRRRMQLEGVGGRARVYNTGLFGAFGRIIQTEGVRGLYRGILPEYYKVVPGVG 584
Query: 196 IGFTVYDTMKSYL 208
I F Y+T+K L
Sbjct: 585 IVFMTYETLKMLL 623
>TC217974 similar to UP|Q9LY28 (Q9LY28) Peroxisomal Ca-dependent solute
carrier-like protein, partial (32%)
Length = 695
Score = 85.9 bits (211), Expect = 1e-17
Identities = 45/135 (33%), Positives = 78/135 (57%), Gaps = 4/135 (2%)
Frame = +2
Query: 78 GIKGIYRGVAPTLFGIFPYAGLKFYFYEEMK----RHVPEDYKKSIMAKLTCGSVAGLLG 133
G + YRG+ P+L G+ PYA + Y+ MK R++ +D + + +L CG+++G +G
Sbjct: 113 GPRAFYRGLVPSLLGMIPYAAIDLTAYDTMKDISKRYILQDSEPGPLVQLGCGTISGAVG 292
Query: 134 QTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPS 193
T Y L+V+R ++Q Q P++ KG + Q +G+ + GL N +KVVP+
Sbjct: 293 ATCVYPLQVIRTRLQAQ--PSNTSDAYKGMFDAFRRTFQLEGFIGFYKGLFPNLLKVVPA 466
Query: 194 AAIGFTVYDTMKSYL 208
A+I + VY+++K L
Sbjct: 467 ASITYVVYESLKKTL 511
Score = 46.6 bits (109), Expect = 9e-06
Identities = 26/97 (26%), Positives = 46/97 (46%)
Frame = +2
Query: 18 ELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEG 77
+L G ++G T V PL+ ++ Q P+ N Y+G+ D +T++
Sbjct: 257 QLGCGTISGAVGATCVYPLQVIRTRLQA-QPS--------NTSDAYKGMFDAFRRTFQLE 409
Query: 78 GIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPED 114
G G Y+G+ P L + P A + + YE +K+ + D
Sbjct: 410 GFIGFYKGLFPNLLKVVPAASITYVVYESLKKTLDLD 520
Score = 30.4 bits (67), Expect = 0.65
Identities = 13/33 (39%), Positives = 21/33 (63%)
Frame = +2
Query: 173 KQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMK 205
++G + + GL + + ++P AAI T YDTMK
Sbjct: 107 QEGPRAFYRGLVPSLLGMIPYAAIDLTAYDTMK 205
>TC215269 similar to PIR|T01729|T01729 mitochondrial solute carrier protein
homolog - Arabidopsis thaliana {Arabidopsis thaliana;} ,
partial (47%)
Length = 809
Score = 85.5 bits (210), Expect = 2e-17
Identities = 54/157 (34%), Positives = 80/157 (50%), Gaps = 19/157 (12%)
Frame = +2
Query: 76 EGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHV--------PEDYKKSIMAKLTCGS 127
E G + +G P++ G+ PY GL F YE +K ++ E+ + S+ +L CG+
Sbjct: 23 EEGPRAYIKGWLPSVIGVIPYVGLNFAVYESLKDYLIKSNPFGLVENSELSVTTRLACGA 202
Query: 128 VAGLLGQTFTYFLEVVRRQMQV--QNLPAS---------EEAELKGTMRSMVLIAQKQGW 176
AG +GQT Y L+V+RR+MQ+ N AS E G + + Q +G+
Sbjct: 203 AAGTVGQTVAYPLDVIRRRMQMVGWNHAASVLTGDGRGKVPLEYTGMVDAFRKTVQHEGF 382
Query: 177 KTLFSGLSINYIKVVPSAAIGFTVYDTMKSYLRVPSR 213
L+ GL N +KVVPS AI F Y+ +K L V R
Sbjct: 383 GALYKGLVPNSVKVVPSIAIAFVTYEVVKDILGVEIR 493
>TC214634 homologue to UP|O49447 (O49447) ADP, ATP carrier-like protein,
partial (83%)
Length = 1503
Score = 79.3 bits (194), Expect = 1e-15
Identities = 54/203 (26%), Positives = 95/203 (46%), Gaps = 7/203 (3%)
Frame = +2
Query: 15 FAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTY 74
F + L GG++ +KT AP+ER+K+L Q + ++ SG ++ + Y+GI DC ++T
Sbjct: 287 FLLDFLMGGVSAAVSKTAAAPIERVKLLIQ--NQDEMIKSGRLS--EPYKGIGDCFTRTM 454
Query: 75 KEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKR-----HVPEDYKKSIMAKLTCGSVA 129
K+ G+ ++RG + FP L F F + KR + Y K L G A
Sbjct: 455 KDEGVIALWRGNTANVIRYFPTQALNFAFKDYFKRLFNFKKDKDGYWKWFAGNLASGGAA 634
Query: 130 GLLGQTFTYFLEVVRRQMQVQNLPASE--EAELKGTMRSMVLIAQKQGWKTLFSGLSINY 187
G F Y L+ R ++ A + E + G + + G L+ G +I+
Sbjct: 635 GASSLLFVYSLDYARTRLANDAKAAKKGGERQFNGLIDVYRKTIKSDGIAGLYRGFNISC 814
Query: 188 IKVVPSAAIGFTVYDTMKSYLRV 210
+ ++ + F +YD++K + V
Sbjct: 815 VGIIVYRGLYFGMYDSLKPVVLV 883
>TC214671 homologue to UP|O49447 (O49447) ADP, ATP carrier-like protein,
partial (85%)
Length = 1562
Score = 79.3 bits (194), Expect = 1e-15
Identities = 54/203 (26%), Positives = 95/203 (46%), Gaps = 7/203 (3%)
Frame = +3
Query: 15 FAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTY 74
F + L GG++ +KT AP+ER+K+L Q + ++ SG ++ + Y+GI DC ++T
Sbjct: 309 FLVDFLMGGVSAAVSKTAAAPIERVKLLIQ--NQDEMIKSGRLS--EPYKGIGDCFARTM 476
Query: 75 KEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKR-----HVPEDYKKSIMAKLTCGSVA 129
K+ G+ ++RG + FP L F F + KR + Y K L G A
Sbjct: 477 KDEGVIALWRGNTANVIRYFPTQALNFAFKDYFKRLFNFKKDKDGYWKWFAGNLASGGAA 656
Query: 130 GLLGQTFTYFLEVVRRQMQVQNLPASE--EAELKGTMRSMVLIAQKQGWKTLFSGLSINY 187
G F Y L+ R ++ A + E + G + + G L+ G +I+
Sbjct: 657 GASSLLFVYSLDYARTRLANDAKAAKKGGERQFNGLVDVYRKTIKSDGVAGLYRGFNISC 836
Query: 188 IKVVPSAAIGFTVYDTMKSYLRV 210
+ ++ + F +YD++K + V
Sbjct: 837 VGIIVYRGLYFGMYDSLKPVVLV 905
>TC208483 similar to GB|AAP21144.1|30102452|BT006336 At5g01500/F7A7_20
{Arabidopsis thaliana;} , partial (54%)
Length = 675
Score = 79.0 bits (193), Expect = 2e-15
Identities = 53/196 (27%), Positives = 88/196 (44%)
Frame = +1
Query: 19 LLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGG 78
L AG AG + + PL+ L++ V E YR + + +E G
Sbjct: 112 LAAGAFAGMTSTFITYPLDVLRLRLAV--------------EPGYRTMSEVALSMLREEG 249
Query: 79 IKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKKSIMAKLTCGSVAGLLGQTFTY 138
Y G+ P+L GI PY + F ++ +K+ +PE Y+K L V+ L Y
Sbjct: 250 FASFYYGLGPSLIGIAPYIAVNFCVFDLLKKSLPEKYQKRTETSLLTAVVSASLATLTCY 429
Query: 139 FLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGF 198
L+ VRRQMQ++ P K + ++ I + G L+ G N +K +P+++I
Sbjct: 430 PLDTVRRQMQLRGTP------YKTVLDAISGIVARDGVIGLYRGFVPNALKNLPNSSIRL 591
Query: 199 TVYDTMKSYLRVPSRD 214
T YD +K + ++
Sbjct: 592 TTYDIVKRLIAASEKE 639
>TC224733 similar to GB|AAP12888.1|30017309|BT006239 At5g48970 {Arabidopsis
thaliana;} , partial (61%)
Length = 1210
Score = 77.4 bits (189), Expect = 5e-15
Identities = 54/197 (27%), Positives = 95/197 (47%), Gaps = 10/197 (5%)
Frame = +2
Query: 21 AGGLAGGFAKTVVAPLERLKILFQV-VSPTK--LNVSGMVNNEQVYRGIRDCLSKTYKEG 77
AG ++GG ++TV +PL+ +KI FQV + PT + + Y G+ +E
Sbjct: 224 AGAISGGISRTVTSPLDVIKIRFQVQLEPTSSWALLRKDLAAASKYTGMFQATKDILREE 403
Query: 78 GIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKKSIMAKLT------CGSVAGL 131
G++G +RG P L + PY ++F ++K K L+ G++AG
Sbjct: 404 GVQGFWRGNVPALLMVMPYTAIQFTVLHKLKTFASGSSKTENHINLSPYLSYISGALAGC 583
Query: 132 LGQTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVL-IAQKQGWKTLFSGLSINYIKV 190
+Y +++R + Q P ++ MRS + I +G++ L+SGLS +++
Sbjct: 584 AATVGSYPFDLLRTILASQGEP-----KVYPNMRSAFMDIVHTRGFQGLYSGLSPTLVEI 748
Query: 191 VPSAAIGFTVYDTMKSY 207
+P A + F YDT K +
Sbjct: 749 IPYAGLQFGTYDTFKRW 799
Score = 45.8 bits (107), Expect = 1e-05
Identities = 33/109 (30%), Positives = 47/109 (42%), Gaps = 1/109 (0%)
Frame = +2
Query: 2 AASSESILDHIPLFAK-ELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNE 60
A+ S +HI L ++G LAG A P + L+ + K
Sbjct: 503 ASGSSKTENHINLSPYLSYISGALAGCAATVGSYPFDLLRTILASQGEPK---------- 652
Query: 61 QVYRGIRDCLSKTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKR 109
VY +R G +G+Y G++PTL I PYAGL+F Y+ KR
Sbjct: 653 -VYPNMRSAFMDIVHTRGFQGLYSGLSPTLVEIIPYAGLQFGTYDTFKR 796
>TC222454 similar to UP|Q9LIF7 (Q9LIF7) Mitochondrial carrier protein-like,
partial (53%)
Length = 743
Score = 77.0 bits (188), Expect = 6e-15
Identities = 47/165 (28%), Positives = 75/165 (44%), Gaps = 17/165 (10%)
Frame = +2
Query: 61 QVYRGIRDCLSKTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHV--------- 111
+VY +R L + G +G+Y G++PTL I PYAGL+F Y+ KR
Sbjct: 68 KVYPNMRAALVDILQTRGFRGLYAGLSPTLVEIIPYAGLQFGTYDTFKRWTMAWNQRQYS 247
Query: 112 -PEDYKKSIMAKLTCGSVAGLLGQTFTYFLEVVRRQMQVQNLPAS-------EEAELKGT 163
P S CG AG + + L+VV+++ Q++ L E K
Sbjct: 248 NPTAESLSSFQLFLCGLAAGTCAKLVCHPLDVVKKRFQIEGLQRHPRYGARVEHRAYKNM 427
Query: 164 MRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYL 208
+ +M I Q +GW L+ G+ + +K P+ A+ F Y+ +L
Sbjct: 428 LDAMKRILQMEGWAGLYKGILPSTVKAAPAGAVTFVAYELTVDWL 562
Score = 40.0 bits (92), Expect = 8e-04
Identities = 24/86 (27%), Positives = 39/86 (44%)
Frame = +2
Query: 20 LAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGI 79
L G AG AK V PL+ +K FQ+ + G + Y+ + D + + + G
Sbjct: 287 LCGLAAGTCAKLVCHPLDVVKKRFQIEGLQRHPRYGARVEHRAYKNMLDAMKRILQMEGW 466
Query: 80 KGIYRGVAPTLFGIFPYAGLKFYFYE 105
G+Y+G+ P+ P + F YE
Sbjct: 467 AGLYKGILPSTVKAAPAGAVTFVAYE 544
>TC212927 similar to UP|Q9ZNY4 (Q9ZNY4) Mitochondrial energy transfer protein
precursor, partial (43%)
Length = 762
Score = 76.6 bits (187), Expect = 8e-15
Identities = 47/164 (28%), Positives = 82/164 (49%), Gaps = 3/164 (1%)
Frame = +2
Query: 48 PTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEM 107
P +L + + VY+ + D + +E G +YRG+ +L G+ PYA + Y+ +
Sbjct: 65 PLELLKTRLTVQRGVYKNLLDAFVRIIQEEGPAELYRGLTSSLIGVVPYAAANYLAYDTL 244
Query: 108 KRHVPEDYKK---SIMAKLTCGSVAGLLGQTFTYFLEVVRRQMQVQNLPASEEAELKGTM 164
++ + +KK + L GS AG + + T+ LEV MQ L + L +
Sbjct: 245 RKAYKKAFKK*EIGNVMTLLIGSAAGAISSSATFPLEVACEHMQAGALNGRQYRNL---L 415
Query: 165 RSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYL 208
++V I +K+G L+ GL ++ +K+VP+A I F Y+ K L
Sbjct: 416 HALVSILEKEGVGGLYRGL*LSCLKLVPAAGISFMCYEACKRVL 547
>TC210946 similar to UP|Q9FI43 (Q9FI43) Calcium-binding transporter-like
protein, partial (42%)
Length = 714
Score = 76.6 bits (187), Expect = 8e-15
Identities = 50/137 (36%), Positives = 75/137 (54%), Gaps = 5/137 (3%)
Frame = +3
Query: 19 LLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKT-YKEG 77
LLAGG+AG A+T + P++ +K Q T SG + + LSK + +
Sbjct: 267 LLAGGIAGAVAQTAIYPMDLVKTRLQ----THACKSGRIPS-------LGTLSKDIWVQE 413
Query: 78 GIKGIYRGVAPTLFGIFPYAGLKFYFYEEMK----RHVPEDYKKSIMAKLTCGSVAGLLG 133
G + YRG+ P+L GI PYAG+ YE +K +++ D + + +L CG+V+G LG
Sbjct: 414 GPRAFYRGLIPSLLGIIPYAGIDLAAYETLKDMSKQYILHDGEPGPLVQLGCGTVSGTLG 593
Query: 134 QTFTYFLEVVRRQMQVQ 150
T Y L+VVR +MQ Q
Sbjct: 594 ATCVYPLQVVRTRMQAQ 644
Score = 69.7 bits (169), Expect = 1e-12
Identities = 46/185 (24%), Positives = 88/185 (46%), Gaps = 7/185 (3%)
Frame = +3
Query: 28 FAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIKGIYRGVA 87
F APL+RLK++ Q+ + I+D +K+GG+ G +RG
Sbjct: 3 FGTRATAPLDRLKVVLQI----------QTTQSHIMPAIKDI----WKKGGLLGFFRGNG 140
Query: 88 PTLFGIFPYAGLKFYFYEEMKRHVP----EDYKKS---IMAKLTCGSVAGLLGQTFTYFL 140
+ + P + ++FY YE +K + E+ K + M +L G +AG + QT Y +
Sbjct: 141 LNVLKVAPESAIRFYSYEMLKTFITRAKGEEAKAANIGAMGRLLAGGIAGAVAQTAIYPM 320
Query: 141 EVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTV 200
++V+ ++Q + L GT+ + + ++G + + GL + + ++P A I
Sbjct: 321 DLVKTRLQTHACKSGRIPSL-GTLSKDIWV--QEGPRAFYRGLIPSLLGIIPYAGIDLAA 491
Query: 201 YDTMK 205
Y+T+K
Sbjct: 492 YETLK 506
Score = 26.9 bits (58), Expect = 7.1
Identities = 16/66 (24%), Positives = 29/66 (43%)
Frame = +3
Query: 18 ELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEG 77
+L G ++G T V PL+ ++ Q ++ Y+G+ D KT +
Sbjct: 558 QLGCGTVSGTLGATCVYPLQVVRTRMQA--------------QRSYKGMADVFRKTLEHE 695
Query: 78 GIKGIY 83
G++G Y
Sbjct: 696 GLRGFY 713
>TC215272 similar to PIR|T01729|T01729 mitochondrial solute carrier protein
homolog - Arabidopsis thaliana {Arabidopsis thaliana;} ,
partial (40%)
Length = 926
Score = 73.6 bits (179), Expect = 7e-14
Identities = 51/145 (35%), Positives = 73/145 (50%), Gaps = 21/145 (14%)
Frame = +3
Query: 90 LFGIF--PYAGLKFYFYEEMKRHV--------PEDYKKSIMAKLTCGSVAGLLGQTFTYF 139
LF +F PY GL F YE +K + +D + S+ +L CG+ AG +GQT Y
Sbjct: 108 LFAVFQIPYVGLNFAVYESLKDWLIKSNPLGLVQDSELSVTTRLACGAAAGTIGQTVAYP 287
Query: 140 LEVVRRQMQV--QNLPASEEA---------ELKGTMRSMVLIAQKQGWKTLFSGLSINYI 188
L+V+RR+MQ+ N AS A G + + + +G+ L+ GL N +
Sbjct: 288 LDVIRRRMQMVGWNHAASVVAGDGRGKVPLAYTGMVDAFRKTVRYEGFGALYRGLVPNSV 467
Query: 189 KVVPSAAIGFTVYDTMKSYLRVPSR 213
KVVPS AI F Y+ +K L V R
Sbjct: 468 KVVPSIAIAFVTYEVVKDILGVEIR 542
>TC214628 homologue to UP|O49875 (O49875) Adenine nucleotide translocator,
complete
Length = 1561
Score = 72.0 bits (175), Expect = 2e-13
Identities = 51/198 (25%), Positives = 89/198 (44%), Gaps = 7/198 (3%)
Frame = +3
Query: 15 FAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTY 74
F + L GG++ +KT AP+ER+K+L Q + ++ +G ++ + Y+GI DC +T
Sbjct: 348 FLLDFLMGGVSAAVSKTAAAPIERVKLLIQ--NQDEMIKTGRLS--EPYKGIGDCFKRTM 515
Query: 75 KEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKR-----HVPEDYKKSIMAKLTCGSVA 129
+ G ++RG + FP L F F + KR + Y K L G A
Sbjct: 516 ADEGAISLWRGNTANVIRYFPTQALNFAFKDYFKRLFNFKKDRDGYWKWFAGNLASGGAA 695
Query: 130 GLLGQTFTYFLEVVRRQMQVQNLPASE--EAELKGTMRSMVLIAQKQGWKTLFSGLSINY 187
G F Y L+ R ++ A + E + G + G L+ G +I+
Sbjct: 696 GASSLLFVYSLDYARTRLANDAKAAKKGGERQFNGLVDVYRKTLASDGVAGLYRGFNISC 875
Query: 188 IKVVPSAAIGFTVYDTMK 205
+ ++ + F +YD++K
Sbjct: 876 VGIIVYRGLYFGLYDSVK 929
>TC224708 similar to GB|AAP12888.1|30017309|BT006239 At5g48970 {Arabidopsis
thaliana;} , partial (52%)
Length = 901
Score = 70.5 bits (171), Expect = 6e-13
Identities = 43/165 (26%), Positives = 73/165 (44%), Gaps = 17/165 (10%)
Frame = +1
Query: 61 QVYRGIRDCLSKTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKR----------H 110
+VY +R G +G+Y G++PTL I PYAGL+F Y+ KR +
Sbjct: 154 KVYPNMRSAFMDIIHTRGFQGLYSGLSPTLVEIIPYAGLQFGTYDTFKRWGMAWNHRYSN 333
Query: 111 VPEDYKKSIMAKLTCGSVAGLLGQTFTYFLEVVRRQMQVQNLPAS-------EEAELKGT 163
+ S CG AG + + L+VV+++ Q++ L E +
Sbjct: 334 TSAEDNLSSFQLFLCGLAAGTCAKLVCHPLDVVKKRFQIEGLQRHPRYGARVEHRAYRNM 513
Query: 164 MRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYL 208
+M I + +GW L+ G+ + +K P+ A+ F Y+ +L
Sbjct: 514 PDAMQRIFRLEGWAGLYKGIIPSTVKAAPAGAVTFVAYELTSDWL 648
Score = 44.3 bits (103), Expect = 4e-05
Identities = 30/102 (29%), Positives = 46/102 (44%)
Frame = +1
Query: 4 SSESILDHIPLFAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVY 63
S+E L LF L G AG AK V PL+ +K FQ+ + G + Y
Sbjct: 337 SAEDNLSSFQLF----LCGLAAGTCAKLVCHPLDVVKKRFQIEGLQRHPRYGARVEHRAY 504
Query: 64 RGIRDCLSKTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYE 105
R + D + + ++ G G+Y+G+ P+ P + F YE
Sbjct: 505 RNMPDAMQRIFRLEGWAGLYKGIIPSTVKAAPAGAVTFVAYE 630
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.138 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,371,511
Number of Sequences: 63676
Number of extensions: 81932
Number of successful extensions: 502
Number of sequences better than 10.0: 118
Number of HSP's better than 10.0 without gapping: 407
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 450
length of query: 217
length of database: 12,639,632
effective HSP length: 93
effective length of query: 124
effective length of database: 6,717,764
effective search space: 833002736
effective search space used: 833002736
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC148360.13


