

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148349.4 - phase: 0
(151 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
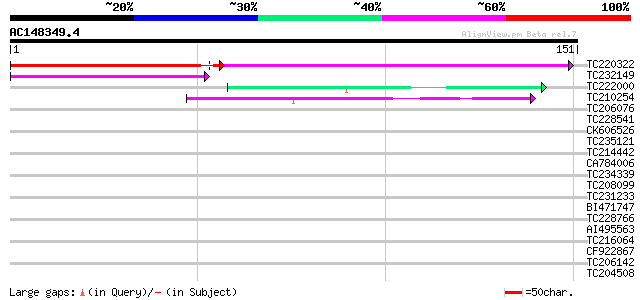
Score E
Sequences producing significant alignments: (bits) Value
TC220322 96 8e-21
TC232149 45 1e-05
TC222000 weakly similar to PIR|H84710|H84710 Mutator-like transp... 44 2e-05
TC210254 similar to GB|AAM26637.1|20855902|AY101516 AT3g06940/F1... 42 1e-04
TC206076 similar to UP|O80698 (O80698) F8K4.12, partial (43%) 35 0.018
TC228541 similar to UP|Q8H4Y5 (Q8H4Y5) Nuclear inhibitor of PP1-... 31 0.20
CK606526 30 0.44
TC235121 similar to GB|AAR92318.1|40823959|BT011282 At5g10310 {A... 30 0.44
TC214442 similar to UP|Q84UB0 (Q84UB0) Transcription factor Myb1... 29 0.75
CA784006 28 1.3
TC234339 similar to UP|Q6J196 (Q6J196) Fasciclin-like AGP 8, par... 28 2.2
TC208099 similar to UP|Q6H6H8 (Q6H6H8) Amino acid permease-like,... 27 2.8
TC231233 similar to GB|BAA95715.1|7939512|AB024028 pectate lyase... 27 2.8
BI471747 27 2.8
TC228766 similar to GB|AAS99672.1|46518381|BT012528 At5g56840 {A... 27 3.7
AI495563 similar to GP|21555010|gb sterol-C-methyltransferase {A... 27 4.8
TC216064 homologue to UP|Q41050 (Q41050) Core protein, partial (... 27 4.8
CF922867 27 4.8
TC206142 similar to UP|Q6U7H9 (Q6U7H9) Pectate lyase, partial (26%) 27 4.8
TC204508 homologue to UP|Q39855 (Q39855) Cyclin, partial (98%) 27 4.8
>TC220322
Length = 1264
Score = 95.5 bits (236), Expect = 8e-21
Identities = 41/97 (42%), Positives = 56/97 (57%)
Frame = +1
Query: 54 CHAWLTLGSYRATYNYFIQPVNSQIYWEPTPYEKSAPPKVRRAAGRPKKNRRKDGNKEPI 113
CH WL++ +Y TY +FI+PV YW T Y PP R GRPKKNR++ +++ +
Sbjct: 538 CHEWLSIEAYNKTYQHFIEPVQGPQYWAQTQYTHPVPPYKRVQRGRPKKNRKRSVDEDNV 717
Query: 114 GRSQMKRTYNDTQCGRCGLIGHNLRGCNKQGVARRPK 150
++KR + CGRCG HN+R C GV RPK
Sbjct: 718 TGHKLKRKLAEFTCGRCGQTNHNIRSCKNIGVPVRPK 828
Score = 59.3 bits (142), Expect = 7e-10
Identities = 27/57 (47%), Positives = 36/57 (62%)
Frame = +2
Query: 1 MRKMSQNKMKLDGRVGPLCPWQQSRLEKEKNASHNWTPLWSGDTPMQRYQIEHCHAW 57
MR M+ K+KL G+ GPLC Q RLEKE + ++ WTP+W GD RY++ H W
Sbjct: 248 MRTMAARKVKLSGKPGPLCLVQYKRLEKEFHFANQWTPIWCGDNMGLRYEV---HMW 409
>TC232149
Length = 1005
Score = 45.4 bits (106), Expect = 1e-05
Identities = 23/53 (43%), Positives = 32/53 (59%)
Frame = -3
Query: 1 MRKMSQNKMKLDGRVGPLCPWQQSRLEKEKNASHNWTPLWSGDTPMQRYQIEH 53
M KM +K+K+ R+ PL P QQSR+EK K S+ T W GD R+++ H
Sbjct: 391 MCKMKSDKLKMATRLRPLVPMQQSRIEKGKVESNK*TANWVGDPDGTRFKVCH 233
>TC222000 weakly similar to PIR|H84710|H84710 Mutator-like transposase
[imported] - Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (12%)
Length = 617
Score = 44.3 bits (103), Expect = 2e-05
Identities = 27/92 (29%), Positives = 36/92 (38%), Gaps = 7/92 (7%)
Frame = +2
Query: 59 TLGSYRATYNYFIQPVNSQIYWEPTPYEKS-------APPKVRRAAGRPKKNRRKDGNKE 111
T S+R TY I P+ ++ W T PPK+RR GRP+K D
Sbjct: 146 TAASFRDTYAETIHPIPGKLEWSKTGNSSMDDNILVVRPPKLRRPPGRPEKRMCVD---- 313
Query: 112 PIGRSQMKRTYNDTQCGRCGLIGHNLRGCNKQ 143
+ R + C RC GH R C +
Sbjct: 314 -----DLNREKHTVHCSRCNQTGHYKRTCKAE 394
>TC210254 similar to GB|AAM26637.1|20855902|AY101516 AT3g06940/F17A9_9
{Arabidopsis thaliana;} , partial (6%)
Length = 814
Score = 42.0 bits (97), Expect = 1e-04
Identities = 29/97 (29%), Positives = 41/97 (41%), Gaps = 4/97 (4%)
Frame = +1
Query: 48 RYQIEHCHAWLTLGSYRATYNYFIQPV----NSQIYWEPTPYEKSAPPKVRRAAGRPKKN 103
R ++C + T+ +YR TY I PV + E T PP +R GRPK
Sbjct: 160 RSPYDYCSRYFTVENYRLTYAESIHPVPNVDKPPVQGESTALVMVIPPPTKRPPGRPKM- 336
Query: 104 RRKDGNKEPIGRSQMKRTYNDTQCGRCGLIGHNLRGC 140
K+ +KR QC +C +GHN + C
Sbjct: 337 ------KQVESIDIIKR---QLQCSKCKGLGHNRKTC 420
>TC206076 similar to UP|O80698 (O80698) F8K4.12, partial (43%)
Length = 1225
Score = 34.7 bits (78), Expect = 0.018
Identities = 11/28 (39%), Positives = 19/28 (67%)
Frame = +1
Query: 72 QPVNSQIYWEPTPYEKSAPPKVRRAAGR 99
+P + ++WEPTP K+ PP R++G+
Sbjct: 157 RPAGANVFWEPTPIAKAPPP*THRSSGK 240
>TC228541 similar to UP|Q8H4Y5 (Q8H4Y5) Nuclear inhibitor of PP1-like,
partial (17%)
Length = 822
Score = 31.2 bits (69), Expect = 0.20
Identities = 16/47 (34%), Positives = 24/47 (51%), Gaps = 3/47 (6%)
Frame = +3
Query: 26 LEKEKNASHNWT---PLWSGDTPMQRYQIEHCHAWLTLGSYRATYNY 69
LEK++N S NW P+W +I C +W+ LG + T N+
Sbjct: 171 LEKDQNPSQNWDL**PIW---------RIIICQSWIFLGIFTCTRNW 284
>CK606526
Length = 513
Score = 30.0 bits (66), Expect = 0.44
Identities = 15/47 (31%), Positives = 22/47 (45%)
Frame = +2
Query: 73 PVNSQIYWEPTPYEKSAPPKVRRAAGRPKKNRRKDGNKEPIGRSQMK 119
P +++ P P K+ P K R P+KN + GN P RS +
Sbjct: 110 PRGKNLWFPPNPK*KTPP*KPGRGGPPPQKNSQPPGNNPPPPRSPQR 250
>TC235121 similar to GB|AAR92318.1|40823959|BT011282 At5g10310 {Arabidopsis
thaliana;} , partial (39%)
Length = 387
Score = 30.0 bits (66), Expect = 0.44
Identities = 13/34 (38%), Positives = 16/34 (46%)
Frame = -1
Query: 26 LEKEKNASHNWTPLWSGDTPMQRYQIEHCHAWLT 59
LEKE +W W +R HCHAW+T
Sbjct: 192 LEKENFHCSSWGKTWVNSFMARRGWNLHCHAWMT 91
>TC214442 similar to UP|Q84UB0 (Q84UB0) Transcription factor Myb1, partial
(30%)
Length = 1102
Score = 29.3 bits (64), Expect = 0.75
Identities = 11/25 (44%), Positives = 14/25 (56%)
Frame = +2
Query: 127 CGRCGLIGHNLRGCNKQGVARRPKD 151
C +CG GHN R C G A P++
Sbjct: 212 CSQCGNNGHNSRTCTDGGAAGSPRE 286
>CA784006
Length = 409
Score = 28.5 bits (62), Expect = 1.3
Identities = 17/60 (28%), Positives = 29/60 (48%), Gaps = 2/60 (3%)
Frame = +1
Query: 1 MRKMSQNKMKLDGRV--GPLCPWQQSRLEKEKNASHNWTPLWSGDTPMQRYQIEHCHAWL 58
+R S ++ +GR+ P+ Q + K++N H P P+ ++I HC AWL
Sbjct: 232 LRSTSSGSLQ-EGRIREDPVSMTSQWLISKKQNLLHQTLP-----NPLLHFRIRHCLAWL 393
>TC234339 similar to UP|Q6J196 (Q6J196) Fasciclin-like AGP 8, partial (22%)
Length = 895
Score = 27.7 bits (60), Expect = 2.2
Identities = 13/31 (41%), Positives = 14/31 (44%)
Frame = -3
Query: 98 GRPKKNRRKDGNKEPIGRSQMKRTYNDTQCG 128
G K RRK PI R + R YN CG
Sbjct: 497 GSSNKLRRKKHLTHPINRKMLIRVYNPIHCG 405
>TC208099 similar to UP|Q6H6H8 (Q6H6H8) Amino acid permease-like, partial
(16%)
Length = 769
Score = 27.3 bits (59), Expect = 2.8
Identities = 9/20 (45%), Positives = 13/20 (65%)
Frame = +3
Query: 114 GRSQMKRTYNDTQCGRCGLI 133
GRS M +N+ QC CG++
Sbjct: 432 GRSSMSHLFNNIQCVHCGMV 491
>TC231233 similar to GB|BAA95715.1|7939512|AB024028 pectate lyase-like
protein {Arabidopsis thaliana;} , partial (34%)
Length = 614
Score = 27.3 bits (59), Expect = 2.8
Identities = 19/57 (33%), Positives = 28/57 (48%), Gaps = 2/57 (3%)
Frame = -1
Query: 6 QNKMKLDGRVGPLCPWQQSRLEKEKNASHNWT--PLWSGDTPMQRYQIEHCHAWLTL 60
Q K L + P W++ + + + H+ T PL S TP + +EH HAWL L
Sbjct: 371 QEKKVLH*DLKP*HSWKRHQHQIA*SMLHSRTCHPLTSNSTP-STHSVEHPHAWLPL 204
>BI471747
Length = 421
Score = 27.3 bits (59), Expect = 2.8
Identities = 13/50 (26%), Positives = 26/50 (52%)
Frame = -2
Query: 71 IQPVNSQIYWEPTPYEKSAPPKVRRAAGRPKKNRRKDGNKEPIGRSQMKR 120
+Q N +I+ P P PP + + +P+K R + G +E G ++ ++
Sbjct: 201 LQRFNGKIHGPPLP-----PPPLSIFSDKPEKQRAEGGERERCGNARRRK 67
>TC228766 similar to GB|AAS99672.1|46518381|BT012528 At5g56840 {Arabidopsis
thaliana;} , partial (54%)
Length = 1220
Score = 26.9 bits (58), Expect = 3.7
Identities = 9/15 (60%), Positives = 10/15 (66%)
Frame = +3
Query: 126 QCGRCGLIGHNLRGC 140
+C CG IGHN R C
Sbjct: 135 KCSHCGTIGHNSRTC 179
>AI495563 similar to GP|21555010|gb sterol-C-methyltransferase {Arabidopsis
thaliana}, partial (31%)
Length = 344
Score = 26.6 bits (57), Expect = 4.8
Identities = 14/60 (23%), Positives = 23/60 (38%), Gaps = 5/60 (8%)
Frame = +3
Query: 76 SQIYWEPTPYEKSAPPKVRRAAGRPKKNRRKDGNKEPIGRSQM-----KRTYNDTQCGRC 130
S ++W P +APP A P+K++ + P + R + CG C
Sbjct: 63 SSVFWAPPSRRANAPPIYPAAPSPPRKSKTTTSSTGPSSLVEPIEDG*DRVLAEPYCGHC 242
>TC216064 homologue to UP|Q41050 (Q41050) Core protein, partial (90%)
Length = 754
Score = 26.6 bits (57), Expect = 4.8
Identities = 11/38 (28%), Positives = 13/38 (33%)
Frame = +2
Query: 20 PWQQSRLEKEKNASHNWTPLWSGDTPMQRYQIEHCHAW 57
PW Q E NW P +R + CH W
Sbjct: 284 PWHQRLEECHDWGCSNWGPGICSQQQQERQDCDRCHYW 397
>CF922867
Length = 450
Score = 26.6 bits (57), Expect = 4.8
Identities = 20/76 (26%), Positives = 28/76 (36%), Gaps = 7/76 (9%)
Frame = +3
Query: 81 EPTPYEKSAPPKVRRAAGRPKKNRRKDGNKEPIGRSQMKRTY-------NDTQCGRCGLI 133
E P + PPK+ + G P + G K+P + K+ + T G I
Sbjct: 84 EKGPPKNPPPPKIAQKRGAPNPKPKTLGKKDPPRGEKGKKAFPPPPGSVGKTPPGAKTGI 263
Query: 134 GHNLRGCNKQGVARRP 149
G R K G R P
Sbjct: 264 GARTRARRKPGPKRAP 311
>TC206142 similar to UP|Q6U7H9 (Q6U7H9) Pectate lyase, partial (26%)
Length = 723
Score = 26.6 bits (57), Expect = 4.8
Identities = 11/33 (33%), Positives = 21/33 (63%)
Frame = -2
Query: 83 TPYEKSAPPKVRRAAGRPKKNRRKDGNKEPIGR 115
T ++ PKV++ + ++N+RK+ KE IG+
Sbjct: 593 TKVDQIRNPKVKKKKKKTRENKRKECRKEIIGK 495
>TC204508 homologue to UP|Q39855 (Q39855) Cyclin, partial (98%)
Length = 1283
Score = 26.6 bits (57), Expect = 4.8
Identities = 13/46 (28%), Positives = 24/46 (51%)
Frame = +2
Query: 72 QPVNSQIYWEPTPYEKSAPPKVRRAAGRPKKNRRKDGNKEPIGRSQ 117
+PV+ ++ +P P EK R K+++K+G+ P +SQ
Sbjct: 47 KPVSKKVIVKPKPSEKVTDIDASPDKKRVLKDKKKEGDANPKKKSQ 184
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.134 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,324,149
Number of Sequences: 63676
Number of extensions: 126238
Number of successful extensions: 940
Number of sequences better than 10.0: 58
Number of HSP's better than 10.0 without gapping: 923
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 936
length of query: 151
length of database: 12,639,632
effective HSP length: 89
effective length of query: 62
effective length of database: 6,972,468
effective search space: 432293016
effective search space used: 432293016
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 54 (25.4 bits)
Medicago: description of AC148349.4


