

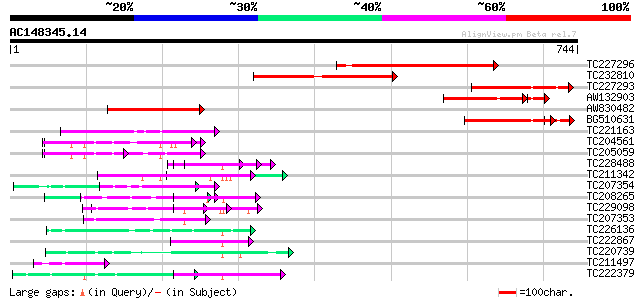
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148345.14 - phase: 0 /pseudo
(744 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC227296 similar to GB|AAN18197.1|23308455|BT000631 At3g05090/T1... 380 e-105
TC232810 similar to GB|AAN18197.1|23308455|BT000631 At3g05090/T1... 327 1e-89
TC227293 similar to GB|AAN18197.1|23308455|BT000631 At3g05090/T1... 232 5e-61
AW132903 similar to GP|22022579|gb AT3g05090/T12H1_5 {Arabidopsi... 207 6e-58
AW830482 similar to GP|22022579|gb AT3g05090/T12H1_5 {Arabidopsi... 216 2e-56
BG510631 213 2e-55
TC221163 67 3e-11
TC204561 homologue to UP|GBLP_SOYBN (Q39836) Guanine nucleotide-... 59 6e-09
TC205059 homologue to UP|GBLP_SOYBN (Q39836) Guanine nucleotide-... 59 6e-09
TC228488 similar to UP|Q8LF96 (Q8LF96) PRL1 protein, partial (70%) 59 1e-08
TC211342 similar to UP|Q9LVF2 (Q9LVF2) Arabidopsis thaliana geno... 57 3e-08
TC207354 weakly similar to UP|WDR5_HUMAN (P61964) WD-repeat prot... 57 3e-08
TC208265 similar to UP|Q6S7B0 (Q6S7B0) TAF5, partial (36%) 57 4e-08
TC229098 similar to PIR|T46032|T46032 WD-40 repeat regulatory pr... 55 9e-08
TC207353 weakly similar to UP|WDR5_HUMAN (P61964) WD-repeat prot... 55 1e-07
TC226136 similar to UP|O48847 (O48847) Expressed protein (At2g32... 55 1e-07
TC222867 similar to UP|Q8L830 (Q8L830) WD40-repeat protein (Frag... 54 2e-07
TC220739 weakly similar to UP|PRP4_HUMAN (O43172) U4/U6 small nu... 53 4e-07
TC211497 weakly similar to UP|Q9FNZ2 (Q9FNZ2) Zfwd1 protein (Fra... 51 2e-06
TC222379 homologue to UP|Q9HGV7 (Q9HGV7) Gbeta like protein, par... 50 4e-06
>TC227296 similar to GB|AAN18197.1|23308455|BT000631 At3g05090/T12H1_5
{Arabidopsis thaliana;} , partial (21%)
Length = 733
Score = 380 bits (976), Expect = e-105
Identities = 183/214 (85%), Positives = 194/214 (90%), Gaps = 1/214 (0%)
Frame = +3
Query: 429 HEVLNNKRHVLTKDTSDSVKLWEITKGVVVEDYGKVSFKEKKEELFEMVSVPGWFTVDTR 488
HEVLNNKRHVLTK LWEITKGVV+EDYGKVSF+EKKEELFEMVS+ WFTVDTR
Sbjct: 3 HEVLNNKRHVLTK-------LWEITKGVVIEDYGKVSFEEKKEELFEMVSISAWFTVDTR 161
Query: 489 LGTLSVHLDTQHCFNAEMYSADLNIVGKPEDDKVNLGRETLKGLLTHWLRKRKQRMGSPA 548
LG+LSVHLDT CF+AEMYSADLN+VGKPEDDKVNL RETLKGLL HWLRKRKQRMGSPA
Sbjct: 162 LGSLSVHLDTPQCFSAEMYSADLNVVGKPEDDKVNLARETLKGLLAHWLRKRKQRMGSPA 341
Query: 549 PANGE-LSGKDVASRSLIHSRAEVDVSSENDATVYPPFEFSVVSPPSIVTEGTHGGPWRK 607
PANGE LSGKD++SRSL HSR EVD SSENDA VYPPFEFSV SPPSI+TEG+HGGPWRK
Sbjct: 342 PANGELLSGKDISSRSLAHSRIEVDGSSENDAMVYPPFEFSVTSPPSIITEGSHGGPWRK 521
Query: 608 KITDLDGTEDEKDFPWWCLDCVLNNRLTPRENTK 641
KITDLDGT+DEKDFPWWCLDCVLNNRL PRENTK
Sbjct: 522 KITDLDGTKDEKDFPWWCLDCVLNNRLPPRENTK 623
>TC232810 similar to GB|AAN18197.1|23308455|BT000631 At3g05090/T12H1_5
{Arabidopsis thaliana;} , partial (24%)
Length = 541
Score = 327 bits (838), Expect = 1e-89
Identities = 162/189 (85%), Positives = 171/189 (89%)
Frame = +1
Query: 320 FSLYLTDLQTRESSLLCTGEHPIRQLALHNDSIWVASTDSSVHRWPAEGCDPQKIFQRGN 379
FSLYLTDLQTRESSLLCTGEHPI QLALH+DSIWVASTDSSVHRWPAEG +PQKIFQRGN
Sbjct: 1 FSLYLTDLQTRESSLLCTGEHPILQLALHDDSIWVASTDSSVHRWPAEGYNPQKIFQRGN 180
Query: 380 SFLAGNLSFSRARVSLEGSTPCF*IMNF**VPIYKEPTLTIRGTPGIVQHEVLNNKRHVL 439
SFLAGNLSFSRARVS+EGSTP VP+YKEPTLTI GTP IVQHEVLNNKRHVL
Sbjct: 181 SFLAGNLSFSRARVSIEGSTP---------VPVYKEPTLTILGTPAIVQHEVLNNKRHVL 333
Query: 440 TKDTSDSVKLWEITKGVVVEDYGKVSFKEKKEELFEMVSVPGWFTVDTRLGTLSVHLDTQ 499
TKD SDSVKLWEITKGVV+EDYGKVSF+EKKEELFEMVS+ WFTVDTRLG+LSVHLDT
Sbjct: 334 TKDASDSVKLWEITKGVVIEDYGKVSFEEKKEELFEMVSISAWFTVDTRLGSLSVHLDTP 513
Query: 500 HCFNAEMYS 508
CF+AEMYS
Sbjct: 514 QCFSAEMYS 540
>TC227293 similar to GB|AAN18197.1|23308455|BT000631 At3g05090/T12H1_5
{Arabidopsis thaliana;} , partial (17%)
Length = 816
Score = 232 bits (591), Expect = 5e-61
Identities = 112/133 (84%), Positives = 121/133 (90%)
Frame = +2
Query: 607 KKITDLDGTEDEKDFPWWCLDCVLNNRLTPRENTKCSFYLQPCEGSSVQILTQGKLSAPR 666
+KITDL+GT+DEKDFPWWCLDCVLNNRL PRENTKCSFYLQPCEGSSVQILTQGKLSAPR
Sbjct: 2 EKITDLNGTKDEKDFPWWCLDCVLNNRLPPRENTKCSFYLQPCEGSSVQILTQGKLSAPR 181
Query: 667 ILRIHKVINYVVEKLVLDNKPLDNLHADGNFAPGVAGSQSLLPVVGDGSFRSGSGLKPWQ 726
ILR+HKVINYV+EK+VLD KPLD+L DG+FAPG+ GSQS L VGDGSFR SG KPWQ
Sbjct: 182 ILRVHKVINYVIEKMVLD-KPLDSLSGDGSFAPGLTGSQSQLQAVGDGSFR--SGFKPWQ 352
Query: 727 KLRPSIEILCCNQ 739
KLRPSIEILC NQ
Sbjct: 353 KLRPSIEILCNNQ 391
>AW132903 similar to GP|22022579|gb AT3g05090/T12H1_5 {Arabidopsis thaliana},
partial (14%)
Length = 422
Score = 207 bits (526), Expect(2) = 6e-58
Identities = 96/110 (87%), Positives = 102/110 (92%)
Frame = +2
Query: 570 EVDVSSENDATVYPPFEFSVVSPPSIVTEGTHGGPWRKKITDLDGTEDEKDFPWWCLDCV 629
EVD SSENDA VYPPFEFSV SPPSI+TEGTHGGPWRKKITDLDGT+DEKDFPWWCLDCV
Sbjct: 11 EVDGSSENDAMVYPPFEFSVTSPPSIITEGTHGGPWRKKITDLDGTKDEKDFPWWCLDCV 190
Query: 630 LNNRLTPRENTKCSFYLQPCEGSSVQILTQGKLSAPRILRIHKVINYVVE 679
LNN L P ENT +FYLQPCEGSS+QILTQGKLSAPRILR+HKVINYV+E
Sbjct: 191 LNNXLPPPENT--NFYLQPCEGSSIQILTQGKLSAPRILRVHKVINYVIE 334
Score = 36.6 bits (83), Expect(2) = 6e-58
Identities = 19/29 (65%), Positives = 23/29 (78%)
Frame = +3
Query: 680 KLVLDNKPLDNLHADGNFAPGVAGSQSLL 708
K+VLD KPLD+L DG+FAP + GSQS L
Sbjct: 333 KMVLD-KPLDSLSGDGSFAPVLTGSQSPL 416
>AW830482 similar to GP|22022579|gb AT3g05090/T12H1_5 {Arabidopsis thaliana},
partial (14%)
Length = 388
Score = 216 bits (551), Expect = 2e-56
Identities = 107/127 (84%), Positives = 117/127 (91%)
Frame = +3
Query: 129 VTCLAAAGKNSNIVASGGLGGEVFVWDIEAALAPVSKCNDAMVDESSNGINGSANLLPST 188
VTCL AA KN+NIVASGGLGGEVF+WDIEAALAPVSKCND VDESSNGINGS NLLP T
Sbjct: 6 VTCLTAAEKNNNIVASGGLGGEVFIWDIEAALAPVSKCNDDTVDESSNGINGSGNLLPLT 185
Query: 189 NLRTISSSHSISLHTAQTQGYIPIAAKGHKESVYALAMDEGGSILVSGGTEKVVRVWDTR 248
+LRTI+SS ++S+HT QTQGYIPIAAKGHK+SVYALAM+E G+ILVSGGTEKVVRVWDTR
Sbjct: 186 SLRTINSSDNMSMHTTQTQGYIPIAAKGHKDSVYALAMNESGTILVSGGTEKVVRVWDTR 365
Query: 249 SGSKILK 255
SGSK LK
Sbjct: 366 SGSKTLK 386
Score = 33.1 bits (74), Expect = 0.46
Identities = 26/121 (21%), Positives = 55/121 (44%), Gaps = 11/121 (9%)
Frame = +3
Query: 225 AMDEGGSILVSGGTEKVVRVWDTRSG-SKILKLKGHTDNIRALLLDSTGRVLFIRIIRFY 283
A ++ +I+ SGG V +WD + + + K T + + ++ +G +L + +R
Sbjct: 21 AAEKNNNIVASGGLGGEVFIWDIEAALAPVSKCNDDTVDESSNGINGSGNLLPLTSLRTI 200
Query: 284 DKGQQRCVHSYAV----------HTDSVWALASTPTFSHVYSGGRDFSLYLTDLQTRESS 333
+ +H+ H DSV+ALA + + + SGG + + + D ++ +
Sbjct: 201 NSSDNMSMHTTQTQGYIPIAAKGHKDSVYALAMNESGTILVSGGTEKVVRVWDTRSGSKT 380
Query: 334 L 334
L
Sbjct: 381 L 383
>BG510631
Length = 431
Score = 213 bits (543), Expect = 2e-55
Identities = 100/119 (84%), Positives = 108/119 (90%)
Frame = +3
Query: 598 EGTHGGPWRKKITDLDGTEDEKDFPWWCLDCVLNNRLTPRENTKCSFYLQPCEGSSVQIL 657
EGTHGGPWRKKITDLDGT+DEKDFPWWCLDCVLNNRL PRENTKCSFYLQPCEGSSVQIL
Sbjct: 3 EGTHGGPWRKKITDLDGTKDEKDFPWWCLDCVLNNRLPPRENTKCSFYLQPCEGSSVQIL 182
Query: 658 TQGKLSAPRILRIHKVINYVVEKLVLDNKPLDNLHADGNFAPGVAGSQSLLPVVGDGSF 716
TQGKLSAPRILRIHKV+NYV+EK+VLD KPLD+L A+G+ APG+ GSQS VGD F
Sbjct: 183 TQGKLSAPRILRIHKVVNYVIEKMVLD-KPLDSLSAEGSIAPGLTGSQSQHQAVGDWIF 356
Score = 48.9 bits (115), Expect = 8e-06
Identities = 25/39 (64%), Positives = 28/39 (71%)
Frame = +1
Query: 703 GSQSLLPVVGDGSFRSGSGLKPWQKLRPSIEILCCNQAS 741
G + +G GSFRSG +PWQKLRPSIEILC NQAS
Sbjct: 316 GHSHSIKQLGIGSFRSG--FQPWQKLRPSIEILCYNQAS 426
>TC221163
Length = 832
Score = 67.0 bits (162), Expect = 3e-11
Identities = 60/212 (28%), Positives = 88/212 (41%), Gaps = 3/212 (1%)
Frame = +3
Query: 67 WALAEDAATCSATFESHVDWVNDAVLVGDNTLVSCSSD-TTLKTWNALSAGTC--VRTHR 123
W D A TF H D V D ++ SD TL+ WN + + VR H
Sbjct: 18 WMWNTDNAALLNTFIGHGDSVTCGDFTPDGKIICTGSDDATLRIWNPKTGESTHVVRGHP 197
Query: 124 QHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALAPVSKCNDAMVDESSNGINGSAN 183
HT+ +TCL S + SG G V + +I N+A+ S +
Sbjct: 198 YHTEGLTCLTI-NSTSTLALSGSKDGSVHIVNITTGRVVD---NNALASHSDS--IECVG 359
Query: 184 LLPSTNLRTISSSHSISLHTAQTQGYIPIAAKGHKESVYALAMDEGGSILVSGGTEKVVR 243
PS + + L + +P H++ V LA G S + SG + VR
Sbjct: 360 FAPSGSWAAVGGMDK-KLIIWDIEHLLPRGTCEHEDGVTCLAW-LGASYVASGCVDGKVR 533
Query: 244 VWDTRSGSKILKLKGHTDNIRALLLDSTGRVL 275
+WD+RSG + LKGH+D I++L + S L
Sbjct: 534 LWDSRSGECVKTLKGHSDAIQSLSVSSNRNYL 629
Score = 37.7 bits (86), Expect = 0.019
Identities = 31/135 (22%), Positives = 57/135 (41%), Gaps = 2/135 (1%)
Frame = +3
Query: 25 VLNDSDDTKHCAGINCLALLTSTISDGSDYLFTGSRDGRLKRWALAE--DAATCSATFES 82
V++++ H I C+ S + G D +L W + TC
Sbjct: 306 VVDNNALASHSDSIECVGFAPS-----GSWAAVGGMDKKLIIWDIEHLLPRGTCE----- 455
Query: 83 HVDWVNDAVLVGDNTLVSCSSDTTLKTWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIV 142
H D V +G + + S D ++ W++ S G CV+T + H+D + L+ + N N +
Sbjct: 456 HEDGVTCLAWLGASYVASGCVDGKVRLWDSRS-GECVKTLKGHSDAIQSLSVS-SNRNYL 629
Query: 143 ASGGLGGEVFVWDIE 157
S + G +++E
Sbjct: 630 VSASVDGTACAFEVE 674
>TC204561 homologue to UP|GBLP_SOYBN (Q39836) Guanine nucleotide-binding
protein beta subunit-like protein, complete
Length = 1419
Score = 59.3 bits (142), Expect = 6e-09
Identities = 61/218 (27%), Positives = 90/218 (40%), Gaps = 7/218 (3%)
Frame = +1
Query: 46 STISDGSDYLFTGSRDGRLKRWALAEDAATCSAT---FESHVDWVNDAVLVGDNTL-VSC 101
+T D SD + T SRD + W L ++ T H +V D VL D +S
Sbjct: 208 ATPIDNSDMIVTASRDKSIILWHLTKEDKTYGVPRRRLTGHSHFVQDVVLSSDGQFALSG 387
Query: 102 SSDTTLKTWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALA 161
S D L+ W+ L+AGT R HT V +A + N IV S + +W+
Sbjct: 388 SWDGELRLWD-LAAGTSARRFVGHTKDVLSVAFSIDNRQIV-SASRDRTIKLWNTLGECK 561
Query: 162 PVSKCNDAMVDESSNGINGSANLLPSTNLRTISSS---HSISLHTAQTQGYIPIAAKGHK 218
+ DA D S PST TI S+ ++ + T + GH
Sbjct: 562 YTIQDGDAHSDWVS-----CVRFSPSTLQPTIVSASWDRTVKVWNL-TNCKLRNTLAGHN 723
Query: 219 ESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKL 256
V +A+ GS+ SGG + V+ +WD G ++ L
Sbjct: 724 GYVNTVAVSPDGSLCASGGKDGVILLWDLAEGKRLYSL 837
Score = 49.7 bits (117), Expect = 5e-06
Identities = 52/223 (23%), Positives = 90/223 (40%), Gaps = 20/223 (8%)
Frame = +1
Query: 43 LLTSTISDGSDYLFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAVLVGDN---TLV 99
+L+ S + + + SRD +K W + ++H DWV+ T+V
Sbjct: 466 VLSVAFSIDNRQIVSASRDRTIKLWNTLGECKYTIQDGDAHSDWVSCVRFSPSTLQPTIV 645
Query: 100 SCSSDTTLKTWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAA 159
S S D T+K WN L+ T H YV +A + + ++ ASGG G + +WD+
Sbjct: 646 SASWDRTVKVWN-LTNCKLRNTLAGHNGYVNTVAVS-PDGSLCASGGKDGVILLWDLAE- 816
Query: 160 LAPVSKCNDAMVDESSNGINGSANLLPSTNLRTISSSHSISLHTAQTQGYI--------- 210
+ + I + P+ ++ SI + +++ +
Sbjct: 817 -------GKRLYSLDAGSIIHALCFSPNRYWLCAATEQSIKIWDLESKSIVEDLKVDLKT 975
Query: 211 -PIAAKG-----HKESVYALAMD--EGGSILVSGGTEKVVRVW 245
A G K+ +Y +++ GS L SG T+ VVRVW
Sbjct: 976 EADATSGGGNANKKKVIYCTSLNWSADGSTLFSGYTDGVVRVW 1104
Score = 35.4 bits (80), Expect = 0.093
Identities = 52/298 (17%), Positives = 94/298 (31%), Gaps = 51/298 (17%)
Frame = +1
Query: 121 THRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALAPVSKCNDAMVDESSNGING 180
T R HTD VT +A NS+++ + + +W + ++ + G+
Sbjct: 172 TMRAHTDVVTAIATPIDNSDMIVTASRDKSIILWHLTK-------------EDKTYGV-- 306
Query: 181 SANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAKGHKESVYALAMDEGGSILVSGGTEK 240
P L GH V + + G +SG +
Sbjct: 307 -----PRRRLT------------------------GHSHFVQDVVLSSDGQFALSGSWDG 399
Query: 241 VVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVL------------------------- 275
+R+WD +G+ + GHT ++ ++ R +
Sbjct: 400 ELRLWDLAAGTSARRFVGHTKDVLSVAFSIDNRQIVSASRDRTIKLWNTLGECKYTIQDG 579
Query: 276 -----FIRIIRFYDKGQQRCV--------------------HSYAVHTDSVWALASTPTF 310
++ +RF Q + ++ A H V +A +P
Sbjct: 580 DAHSDWVSCVRFSPSTLQPTIVSASWDRTVKVWNLTNCKLRNTLAGHNGYVNTVAVSPDG 759
Query: 311 SHVYSGGRDFSLYLTDLQTRESSLLCTGEHPIRQLALHNDSIWV-ASTDSSVHRWPAE 367
S SGG+D + L DL + I L + W+ A+T+ S+ W E
Sbjct: 760 SLCASGGKDGVILLWDLAEGKRLYSLDAGSIIHALCFSPNRYWLCAATEQSIKIWDLE 933
>TC205059 homologue to UP|GBLP_SOYBN (Q39836) Guanine nucleotide-binding
protein beta subunit-like protein, complete
Length = 1492
Score = 59.3 bits (142), Expect = 6e-09
Identities = 61/218 (27%), Positives = 90/218 (40%), Gaps = 7/218 (3%)
Frame = +2
Query: 46 STISDGSDYLFTGSRDGRLKRWALAEDAATCSAT---FESHVDWVNDAVLVGDNTL-VSC 101
+T D SD + T SRD + W L ++ T H +V D VL D +S
Sbjct: 284 ATPIDNSDMIVTASRDKSIILWHLTKEDKTYGVPRRRLTGHSHFVQDVVLSSDGQFALSG 463
Query: 102 SSDTTLKTWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALA 161
S D L+ W+ L+AGT R HT V +A + N IV S + +W+
Sbjct: 464 SWDGELRLWD-LAAGTSARRFVGHTKDVLSVAFSIDNRQIV-SASRDRTIKLWNTLGECK 637
Query: 162 PVSKCNDAMVDESSNGINGSANLLPSTNLRTISSS---HSISLHTAQTQGYIPIAAKGHK 218
+ DA D S PST TI S+ ++ + T + GH
Sbjct: 638 YTIQDGDAHSDWVS-----CVRFSPSTLQPTIVSASWDRTVKVWNL-TNCKLRNTLAGHN 799
Query: 219 ESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKL 256
V +A+ GS+ SGG + V+ +WD G ++ L
Sbjct: 800 GYVNTVAVSPDGSLCASGGKDGVILLWDLAEGKRLYSL 913
Score = 47.8 bits (112), Expect = 2e-05
Identities = 33/117 (28%), Positives = 54/117 (45%), Gaps = 3/117 (2%)
Frame = +2
Query: 43 LLTSTISDGSDYLFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAVLVGDN---TLV 99
+L+ S + + + SRD +K W + ++H DWV+ T+V
Sbjct: 542 VLSVAFSIDNRQIVSASRDRTIKLWNTLGECKYTIQDGDAHSDWVSCVRFSPSTLQPTIV 721
Query: 100 SCSSDTTLKTWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDI 156
S S D T+K WN L+ T H YV +A + + ++ ASGG G + +WD+
Sbjct: 722 SASWDRTVKVWN-LTNCKLRNTLAGHNGYVNTVAVS-PDGSLCASGGKDGVILLWDL 886
Score = 35.4 bits (80), Expect = 0.093
Identities = 52/298 (17%), Positives = 94/298 (31%), Gaps = 51/298 (17%)
Frame = +2
Query: 121 THRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALAPVSKCNDAMVDESSNGING 180
T R HTD VT +A NS+++ + + +W + ++ + G+
Sbjct: 248 TMRAHTDVVTAIATPIDNSDMIVTASRDKSIILWHLTK-------------EDKTYGV-- 382
Query: 181 SANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAKGHKESVYALAMDEGGSILVSGGTEK 240
P L GH V + + G +SG +
Sbjct: 383 -----PRRRLT------------------------GHSHFVQDVVLSSDGQFALSGSWDG 475
Query: 241 VVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVL------------------------- 275
+R+WD +G+ + GHT ++ ++ R +
Sbjct: 476 ELRLWDLAAGTSARRFVGHTKDVLSVAFSIDNRQIVSASRDRTIKLWNTLGECKYTIQDG 655
Query: 276 -----FIRIIRFYDKGQQRCV--------------------HSYAVHTDSVWALASTPTF 310
++ +RF Q + ++ A H V +A +P
Sbjct: 656 DAHSDWVSCVRFSPSTLQPTIVSASWDRTVKVWNLTNCKLRNTLAGHNGYVNTVAVSPDG 835
Query: 311 SHVYSGGRDFSLYLTDLQTRESSLLCTGEHPIRQLALHNDSIWV-ASTDSSVHRWPAE 367
S SGG+D + L DL + I L + W+ A+T+ S+ W E
Sbjct: 836 SLCASGGKDGVILLWDLAEGKRLYSLDAGSIIHALCFSPNRYWLCAATEQSIKIWDLE 1009
>TC228488 similar to UP|Q8LF96 (Q8LF96) PRL1 protein, partial (70%)
Length = 1303
Score = 58.5 bits (140), Expect = 1e-08
Identities = 38/146 (26%), Positives = 66/146 (45%), Gaps = 5/146 (3%)
Frame = +3
Query: 208 GYIPIAAKGHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALL 267
G + + GH E V LA+ + + S G +K V+ WD I GH + L
Sbjct: 606 GVLKLTLTGHIEQVRGLAVSNRHTYMFSAGDDKQVKCWDLEQNKVIRSYHGHLSGVYCLA 785
Query: 268 LDSTGRVLFI----RIIRFYDKGQQRCVHSYAVHTDSVWALASTPTFSHVYSGGRDFSLY 323
L T VL + R +D + +H+ + H ++V ++ + PT V +G D ++
Sbjct: 786 LHPTIDVLLTGGRDSVCRVWDIRSKMQIHALSGHDNTVCSVFTRPTDPQVVTGSHDTTIK 965
Query: 324 LTDLQ-TRESSLLCTGEHPIRQLALH 348
+ DL+ + S L + +R +A H
Sbjct: 966 MWDLRYGKTMSTLTNHKKSVRAMAQH 1043
Score = 52.8 bits (125), Expect = 6e-07
Identities = 28/105 (26%), Positives = 52/105 (48%), Gaps = 4/105 (3%)
Frame = +3
Query: 230 GSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVLFI----RIIRFYDK 285
G +L + +++WD SG L L GH + +R L + + +F + ++ +D
Sbjct: 546 GHVLYGMHHGRTIKIWDLASGVLKLTLTGHIEQVRGLAVSNRHTYMFSAGDDKQVKCWDL 725
Query: 286 GQQRCVHSYAVHTDSVWALASTPTFSHVYSGGRDFSLYLTDLQTR 330
Q + + SY H V+ LA PT + +GGRD + D++++
Sbjct: 726 EQNKVIRSYHGHLSGVYCLALHPTIDVLLTGGRDSVCRVWDIRSK 860
Score = 47.4 bits (111), Expect = 2e-05
Identities = 29/97 (29%), Positives = 46/97 (46%), Gaps = 4/97 (4%)
Frame = +3
Query: 216 GHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVL 275
GH VY LA+ +L++GG + V RVWD RS +I L GH + + ++ T +
Sbjct: 756 GHLSGVYCLALHPTIDVLLTGGRDSVCRVWDIRSKMQIHALSGHDNTVCSVFTRPTDPQV 935
Query: 276 FI----RIIRFYDKGQQRCVHSYAVHTDSVWALASTP 308
I+ +D + + + H SV A+A P
Sbjct: 936 VTGSHDTTIKMWDLRYGKTMSTLTNHKKSVRAMAQHP 1046
Score = 32.0 bits (71), Expect = 1.0
Identities = 43/210 (20%), Positives = 81/210 (38%), Gaps = 7/210 (3%)
Frame = +3
Query: 48 ISDGSDYLFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAVLVGD-NTLVSCSSDTT 106
+S+ Y+F+ D ++K W L ++ S + H+ V L + L++ D+
Sbjct: 660 VSNRHTYMFSAGDDKQVKCWDLEQNKVIRS--YHGHLSGVYCLALHPTIDVLLTGGRDSV 833
Query: 107 LKTWNALSAGTCVRTHR-QHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIE-----AAL 160
+ W+ S ++ H D C V +G + +WD+ + L
Sbjct: 834 CRVWDIRSK---MQIHALSGHDNTVCSVFTRPTDPQVVTGSHDTTIKMWDLRYGKTMSTL 1004
Query: 161 APVSKCNDAMVDESSNGINGSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAKGHKES 220
K AM SA+ + N++ + H +Q K
Sbjct: 1005TNHKKSVRAMAQHPKEQAFASAS---ADNIKKFNLPKGEFCHNMLSQ---------QKTI 1148
Query: 221 VYALAMDEGGSILVSGGTEKVVRVWDTRSG 250
+ A+A++E G ++V+GG + WD +SG
Sbjct: 1149INAMAVNEEG-VMVTGGDNGSMWFWDWKSG 1235
Score = 31.6 bits (70), Expect = 1.3
Identities = 29/101 (28%), Positives = 43/101 (41%), Gaps = 1/101 (0%)
Frame = +3
Query: 34 HCAGINCLALLTSTISDGSDYLFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAVL- 92
H +G+ CLAL + D L TG RD + W + + + H + V
Sbjct: 759 HLSGVYCLALHPTI-----DVLLTGGRDSVCRVWDIRSKMQIHALS--GHDNTVCSVFTR 917
Query: 93 VGDNTLVSCSSDTTLKTWNALSAGTCVRTHRQHTDYVTCLA 133
D +V+ S DTT+K W+ L G + T H V +A
Sbjct: 918 PTDPQVVTGSHDTTIKMWD-LRYGKTMSTLTNHKKSVRAMA 1037
>TC211342 similar to UP|Q9LVF2 (Q9LVF2) Arabidopsis thaliana genomic DNA,
chromosome 3, P1 clone: MIL23, partial (29%)
Length = 862
Score = 57.0 bits (136), Expect = 3e-08
Identities = 49/223 (21%), Positives = 98/223 (42%), Gaps = 16/223 (7%)
Frame = +1
Query: 116 GTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDI--EAALAPVSKCNDAMVDE 173
GTC T H VT L K +++ASG +V +WD+ E L + D +
Sbjct: 10 GTCETTLNGHKGAVTTLRY-NKAGSLLASGSRDNDVILWDVVGETGLFRLRGHRDQAAKQ 186
Query: 174 ------SSNGINGSANLLP-STNLRTISSS---HSISLHTAQTQGYIPIAAKGHKESVYA 223
S+ +N A ++ S + + I+ + ++ +H A T + ++ GHK V
Sbjct: 187 LTVSNVSTMKMNDDALVVAISPDAKYIAVALLDSTVKVHFADTFKFF-LSLYGHKLPVLC 363
Query: 224 LAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVLFI----RI 279
+ + G ++V+G +K +++W G + H D++ A+ +F R+
Sbjct: 364 MDISSDGDLIVTGSADKNIKIWGLDFGDCHKSIFAHADSVMAVQFVPKTHYVFSVGKDRL 543
Query: 280 IRFYDKGQQRCVHSYAVHTDSVWALASTPTFSHVYSGGRDFSL 322
++++D + + + H +W LA + + +G D S+
Sbjct: 544 VKYWDADKFELLLTLEGHHADIWCLAVSNRGDFIVTGSHDRSI 672
Score = 52.4 bits (124), Expect = 7e-07
Identities = 57/225 (25%), Positives = 79/225 (34%), Gaps = 67/225 (29%)
Frame = +1
Query: 207 QGYIPIAAKGHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDN---- 262
+G GHK +V L ++ GS+L SG + V +WD + + +L+GH D
Sbjct: 7 KGTCETTLNGHKGAVTTLRYNKAGSLLASGSRDNDVILWDVVGETGLFRLRGHRDQAAKQ 186
Query: 263 --------------------------IRALLLDSTGRVLFIRIIRFY------------- 283
I LLDST +V F +F+
Sbjct: 187 LTVSNVSTMKMNDDALVVAISPDAKYIAVALLDSTVKVHFADTFKFFLSLYGHKLPVLCM 366
Query: 284 --------------DKGQQ-------RCVHSYAVHTDSVWALASTPTFSHVYSGGRDFSL 322
DK + C S H DSV A+ P +V+S G+D +
Sbjct: 367 DISSDGDLIVTGSADKNIKIWGLDFGDCHKSIFAHADSVMAVQFVPKTHYVFSVGKDRLV 546
Query: 323 YLTDLQTRESSLLCTGEH-PIRQLALHN--DSIWVASTDSSVHRW 364
D E L G H I LA+ N D I S D S+ RW
Sbjct: 547 KYWDADKFELLLTLEGHHADIWCLAVSNRGDFIVTGSHDRSIRRW 681
Score = 41.2 bits (95), Expect = 0.002
Identities = 34/116 (29%), Positives = 47/116 (40%), Gaps = 1/116 (0%)
Frame = +1
Query: 41 LALLTSTISDGSDYLFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAVLVGDNTLV- 99
L +L IS D + TGS D +K W L D C + +H D V V V
Sbjct: 349 LPVLCMDISSDGDLIVTGSADKNIKIWGL--DFGDCHKSIFAHADSVMAVQFVPKTHYVF 522
Query: 100 SCSSDTTLKTWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWD 155
S D +K W+A + H D + CLA + + IV +G + WD
Sbjct: 523 SVGKDRLVKYWDADKFELLLTLEGHHAD-IWCLAVSNRGDFIV-TGSHDRSIRRWD 684
Score = 30.4 bits (67), Expect = 3.0
Identities = 10/35 (28%), Positives = 20/35 (56%)
Frame = +1
Query: 212 IAAKGHKESVYALAMDEGGSILVSGGTEKVVRVWD 246
+ +GH ++ LA+ G +V+G ++ +R WD
Sbjct: 580 LTLEGHHADIWCLAVSNRGDFIVTGSHDRSIRRWD 684
Score = 30.0 bits (66), Expect = 3.9
Identities = 15/39 (38%), Positives = 22/39 (55%)
Frame = +1
Query: 34 HCAGINCLALLTSTISDGSDYLFTGSRDGRLKRWALAED 72
H A I CLA +S+ D++ TGS D ++RW E+
Sbjct: 595 HHADIWCLA-----VSNRGDFIVTGSHDRSIRRWDRTEE 696
>TC207354 weakly similar to UP|WDR5_HUMAN (P61964) WD-repeat protein 5,
partial (57%)
Length = 1124
Score = 57.0 bits (136), Expect = 3e-08
Identities = 58/248 (23%), Positives = 99/248 (39%), Gaps = 3/248 (1%)
Frame = +1
Query: 6 STGNANNSTRPRKEKRLTYVLNDSDDTKHCAGINCLALLTSTISDGSDYLFTGSRDGRLK 65
S+G T+ K R L D ++ C + +DG+ L + S D L
Sbjct: 64 SSGGGGTGTQTFKPYRHLKTLTDHENAVSCVKFS---------NDGT-LLASASLDKTLI 213
Query: 66 RWALAEDAATCSATFESHVDWVNDAVLVGDNTLV-SCSSDTTLKTWNALSAGTCVRTHRQ 124
W+ A T H + ++D D+ + S S D TL+ W+A G C++ R
Sbjct: 214 IWSSA--TLTLCHRLVGHSEGISDLAWSSDSHYICSASDDRTLRIWDATVGGGCIKILRG 387
Query: 125 HTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALAPVSKCNDAMVDESSNGINGSANL 184
H D V C+ ++S IV SG + VWD++ V + +S N NL
Sbjct: 388 HDDAVFCVNFNPQSSYIV-SGSFDETIKVWDVKTGKC-VHTIKGHTMPVTSVHYNRDGNL 561
Query: 185 LPSTNLRTISSSH--SISLHTAQTQGYIPIAAKGHKESVYALAMDEGGSILVSGGTEKVV 242
+ IS+SH S + +T + + +V G ++++ +
Sbjct: 562 I-------ISASHDGSCKIWDTETGNLLKTLIEDKAPAVSFAKFSPNGKLILAATLNDTL 720
Query: 243 RVWDTRSG 250
++W+ SG
Sbjct: 721 KLWNYGSG 744
Score = 52.8 bits (125), Expect = 6e-07
Identities = 34/157 (21%), Positives = 77/157 (48%)
Frame = +1
Query: 119 VRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALAPVSKCNDAMVDESSNGI 178
++T H + V+C+ + + ++AS L + +W + A ++ C+ + S GI
Sbjct: 115 LKTLTDHENAVSCVKFSN-DGTLLASASLDKTLIIW----SSATLTLCHRLV--GHSEGI 273
Query: 179 NGSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAKGHKESVYALAMDEGGSILVSGGT 238
+ A S + + S ++ + A G +GH ++V+ + + S +VSG
Sbjct: 274 SDLAWSSDSHYICSASDDRTLRIWDATVGGGCIKILRGHDDAVFCVNFNPQSSYIVSGSF 453
Query: 239 EKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVL 275
++ ++VWD ++G + +KGHT + ++ + G ++
Sbjct: 454 DETIKVWDVKTGKCVHTIKGHTMPVTSVHYNRDGNLI 564
Score = 33.1 bits (74), Expect = 0.46
Identities = 14/58 (24%), Positives = 29/58 (49%), Gaps = 2/58 (3%)
Frame = +2
Query: 105 TTLKTWNALSAGTCVRTHRQHTDYVTCLAA--AGKNSNIVASGGLGGEVFVWDIEAAL 160
TTL ++ + G C++ + H + V C+ + + N + G V++WD++ L
Sbjct: 710 TTLLSYGTMGVGKCLKIYSGHVNRVYCITSTFSVTNGKYIVGGSEDHCVYIWDLQQKL 883
>TC208265 similar to UP|Q6S7B0 (Q6S7B0) TAF5, partial (36%)
Length = 1048
Score = 56.6 bits (135), Expect = 4e-08
Identities = 34/118 (28%), Positives = 54/118 (44%), Gaps = 4/118 (3%)
Frame = +2
Query: 216 GHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVL 275
GH V + + + +G ++K VR+WD +SG + GH I +L + GR +
Sbjct: 233 GHLSDVDCVQWHANCNYIATGSSDKTVRLWDVQSGECVRVFVGHRGMILSLAMSPDGRYM 412
Query: 276 FIR----IIRFYDKGQQRCVHSYAVHTDSVWALASTPTFSHVYSGGRDFSLYLTDLQT 329
I +D RC+ HT VW+LA + S + SG D ++ L D+ T
Sbjct: 413 ASGDEDGTIMMWDLSSGRCLTPLIGHTSCVWSLAFSSEGSVIASGSADCTVKLWDVNT 586
Score = 51.6 bits (122), Expect = 1e-06
Identities = 47/224 (20%), Positives = 90/224 (39%), Gaps = 3/224 (1%)
Frame = +2
Query: 46 STISDGSDYLFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAVLVGDNTLVSCSSDT 105
++ S D++ + S D ++ W+ +A ++ W VG + S S D
Sbjct: 5 ASFSPVGDFILSSSADSTIRLWSTKLNANLVCYKGHNYPVWDVQFSPVG-HYFASSSHDR 181
Query: 106 TLKTWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALAPVSK 165
T + W+ + +R H V C+ N N +A+G V +WD+++ +
Sbjct: 182 TARIWS-MDRIQPLRIMAGHLSDVDCVQWHA-NCNYIATGSSDKTVRLWDVQSG-----E 340
Query: 166 CNDAMVDESSNGINGSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAKGHKESVYALA 225
C V G+ S + P + + G GH V++LA
Sbjct: 341 CVRVFVGH--RGMILSLAMSPDGRYMASGDEDGTIMMWDLSSGRCLTPLIGHTSCVWSLA 514
Query: 226 MDEGGSILVSGGTEKVVRVWDTRSGSKILKLK---GHTDNIRAL 266
GS++ SG + V++WD + +K+ + + G + +R+L
Sbjct: 515 FSSEGSVIASGSADCTVKLWDVNTSTKVSRAEEKGGSANRLRSL 646
Score = 46.2 bits (108), Expect = 5e-05
Identities = 44/185 (23%), Positives = 80/185 (42%), Gaps = 2/185 (1%)
Frame = +2
Query: 93 VGDNTLVSCSSDTTLKTWNA-LSAG-TCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGE 150
VGD ++S S+D+T++ W+ L+A C + H +Y + AS
Sbjct: 20 VGD-FILSSSADSTIRLWSTKLNANLVCYKGH----NYPVWDVQFSPVGHYFASSSHDRT 184
Query: 151 VFVWDIEAALAPVSKCNDAMVDESSNGINGSANLLPSTNLRTISSSHSISLHTAQTQGYI 210
+W ++ + P+ + D + + N + T SS ++ L Q+ +
Sbjct: 185 ARIWSMDR-IQPLRIMAGHLSDVDCVQWHANCNYIA-----TGSSDKTVRLWDVQSGECV 346
Query: 211 PIAAKGHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDS 270
+ GH+ + +LAM G + SG + + +WD SG + L GHT + +L S
Sbjct: 347 RVFV-GHRGMILSLAMSPDGRYMASGDEDGTIMMWDLSSGRCLTPLIGHTSCVWSLAFSS 523
Query: 271 TGRVL 275
G V+
Sbjct: 524 EGSVI 538
>TC229098 similar to PIR|T46032|T46032 WD-40 repeat regulatory protein tup1
homolog - Arabidopsis thaliana {Arabidopsis thaliana;} ,
partial (59%)
Length = 580
Score = 55.5 bits (132), Expect = 9e-08
Identities = 40/170 (23%), Positives = 77/170 (44%), Gaps = 4/170 (2%)
Frame = +3
Query: 96 NTLVSCSSDTTLKTWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWD 155
N +VS S D T++ W+ S G C++ H+D VT + ++ +++ S G +WD
Sbjct: 93 NIIVSGSFDETVRVWDVKS-GKCLKVLPAHSDPVTAVDF-NRDGSLIVSSSYDGLCRIWD 266
Query: 156 IEAALAPVSKCNDAMVDESSNGINGSANLLPSTNLRTISS-SHSISLHTAQTQGYIPIAA 214
A C ++D+ + ++ P+ + + +++ L T ++
Sbjct: 267 -----ASTGHCMKTLIDDDNPPVS-FVKFSPNAKFILVGTLDNTLRLWNYSTGKFLKTYT 428
Query: 215 KGHKESVYALAMD---EGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTD 261
GH S Y ++ G +V G E + +WD +S + KL+GH+D
Sbjct: 429 -GHVNSKYCISSTFSTTNGKYIVGGSEENYIYLWDLQSRKIVQKLEGHSD 575
Score = 51.6 bits (122), Expect = 1e-06
Identities = 50/233 (21%), Positives = 95/233 (40%), Gaps = 9/233 (3%)
Frame = +3
Query: 108 KTWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALAPVSKCN 167
+ W+ + G+ ++T HT+YV C+ SNI+ SG V VWD+++ KC
Sbjct: 3 RLWD-VPTGSLIKTLHGHTNYVFCVNF-NPQSNIIVSGSFDETVRVWDVKSG-----KC- 158
Query: 168 DAMVDESSNGINGSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAKGHKESVYALAMD 227
+LP+ H + V A+ +
Sbjct: 159 --------------LKVLPA-----------------------------HSDPVTAVDFN 209
Query: 228 EGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVLFIRI------IR 281
GS++VS + + R+WD +G +K DN + + FI + +R
Sbjct: 210 RDGSLIVSSSYDGLCRIWDASTGH-CMKTLIDDDNPPVSFVKFSPNAKFILVGTLDNTLR 386
Query: 282 FYDKGQQRCVHSYAVHTDSVWALASTPTFS---HVYSGGRDFSLYLTDLQTRE 331
++ + + +Y H +S + ++ST + + ++ G + +YL DLQ+R+
Sbjct: 387 LWNYSTGKFLKTYTGHVNSKYCISSTFSTTNGKYIVGGSEENYIYLWDLQSRK 545
Score = 44.3 bits (103), Expect = 2e-04
Identities = 22/80 (27%), Positives = 41/80 (50%), Gaps = 4/80 (5%)
Frame = +3
Query: 216 GHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVL 275
GH V+ + + +I+VSG ++ VRVWD +SG + L H+D + A+ + G ++
Sbjct: 48 GHTNYVFCVNFNPQSNIIVSGSFDETVRVWDVKSGKCLKVLPAHSDPVTAVDFNRDGSLI 227
Query: 276 ----FIRIIRFYDKGQQRCV 291
+ + R +D C+
Sbjct: 228 VSSSYDGLCRIWDASTGHCM 287
Score = 36.2 bits (82), Expect = 0.054
Identities = 29/130 (22%), Positives = 55/130 (42%), Gaps = 8/130 (6%)
Frame = +3
Query: 243 RVWDTRSGSKILKLKGHTDNIRALLLDSTGRVL----FIRIIRFYDKGQQRCVHSYAVHT 298
R+WD +GS I L GHT+ + + + ++ F +R +D +C+ H+
Sbjct: 3 RLWDVPTGSLIKTLHGHTNYVFCVNFNPQSNIIVSGSFDETVRVWDVKSGKCLKVLPAHS 182
Query: 299 DSVWALASTPTFSHVYSGGRDFSLYLTDLQTRE--SSLLCTGEHPIRQLALHNDS--IWV 354
D V A+ S + S D + D T +L+ P+ + ++ I V
Sbjct: 183 DPVTAVDFNRDGSLIVSSSYDGLCRIWDASTGHCMKTLIDDDNPPVSFVKFSPNAKFILV 362
Query: 355 ASTDSSVHRW 364
+ D+++ W
Sbjct: 363 GTLDNTLRLW 392
>TC207353 weakly similar to UP|WDR5_HUMAN (P61964) WD-repeat protein 5,
partial (58%)
Length = 898
Score = 54.7 bits (130), Expect = 1e-07
Identities = 45/171 (26%), Positives = 76/171 (44%), Gaps = 5/171 (2%)
Frame = +3
Query: 98 LVSCSSDTTLKTWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIE 157
+VS S D T+K W+ + G CV T + HT VT + ++ ++ S G +WD
Sbjct: 39 IVSGSFDETIKVWD-VKTGKCVHTIKGHTMPVTSVHY-NRDGTLIISASHDGSCKIWDTG 212
Query: 158 AA--LAPVSKCNDAMVDESSNGINGSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAK 215
L + + V + NG L + N ++ L + ++ I +
Sbjct: 213 TGNLLKTLIEDKAPAVSFAKFSPNGKFILAATLN-------DTLKLWNYGSGKFLKIYS- 368
Query: 216 GHKESVYALAMD---EGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNI 263
GH VY + G +VSG ++ V +WD ++ + I KL+GHTD +
Sbjct: 369 GHVNRVYCITSTFSVTNGRYIVSGSEDRCVYIWDLQAKNMIQKLEGHTDTV 521
Score = 40.0 bits (92), Expect = 0.004
Identities = 34/182 (18%), Positives = 73/182 (39%), Gaps = 50/182 (27%)
Frame = +3
Query: 221 VYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHT-------------------- 260
V+ + + S +VSG ++ ++VWD ++G + +KGHT
Sbjct: 3 VFCVNFNPQSSYIVSGSFDETIKVWDVKTGKCVHTIKGHTMPVTSVHYNRDGTLIISASH 182
Query: 261 ------------DNIRALLLDSTGRVLFIRI---------------IRFYDKGQQRCVHS 293
+ ++ L+ D V F + ++ ++ G + +
Sbjct: 183 DGSCKIWDTGTGNLLKTLIEDKAPAVSFAKFSPNGKFILAATLNDTLKLWNYGSGKFLKI 362
Query: 294 YAVHTDSVWALASTPTFS---HVYSGGRDFSLYLTDLQTRESSLLCTGEHPIRQLALHND 350
Y+ H + V+ + ST + + ++ SG D +Y+ DLQ + + I++L H D
Sbjct: 363 YSGHVNRVYCITSTFSVTNGRYIVSGSEDRCVYIWDLQAK---------NMIQKLEGHTD 515
Query: 351 SI 352
++
Sbjct: 516 TV 521
>TC226136 similar to UP|O48847 (O48847) Expressed protein
(At2g32700/F24L7.16), partial (42%)
Length = 1463
Score = 54.7 bits (130), Expect = 1e-07
Identities = 61/279 (21%), Positives = 111/279 (38%), Gaps = 5/279 (1%)
Frame = +3
Query: 49 SDGSDYLFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAVLVGDNTLVSCSS-DTTL 107
SDG L + D ++ W + + +T E H + D ++T ++ SS DTT+
Sbjct: 261 SDGK-LLASAGHDKKVVLWNM--ETLQTESTPEEHSLIITDVRFRPNSTQLATSSFDTTV 431
Query: 108 KTWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALAPVSKCN 167
+ W+A + T+ HT +V L K + + S E+ W I +
Sbjct: 432 RLWDAADPTFPLHTYSGHTSHVVSLDFHPKKTELFCSCDNNNEIRFWSIS------QYSS 593
Query: 168 DAMVDESSNGINGSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAKGHKESVYALAMD 227
+ S + L +L +S + +SL +T + +GH V+ + D
Sbjct: 594 TRVFKGGSTQVRFQPRL---GHLLAAASGNVVSLFDVETDRQMH-TLQGHSAEVHCVCWD 761
Query: 228 EGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVLFI----RIIRFY 283
G L S E V+VW SG I +L + + + + L + + + +
Sbjct: 762 TNGDYLASVSQES-VKVWSLASGECIHELNSSGNMFHSCVFHPSYSTLLVIGGYQSLELW 938
Query: 284 DKGQQRCVHSYAVHTDSVWALASTPTFSHVYSGGRDFSL 322
+ + +C+ + H + ALA +P V S D S+
Sbjct: 939 NMAENKCM-TIPAHECVISALAQSPLTGMVASASHDKSV 1052
>TC222867 similar to UP|Q8L830 (Q8L830) WD40-repeat protein (Fragment),
partial (23%)
Length = 591
Score = 54.3 bits (129), Expect = 2e-07
Identities = 28/112 (25%), Positives = 51/112 (45%), Gaps = 4/112 (3%)
Frame = +1
Query: 212 IAAKGHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDST 271
+ KGHK ++++ +V+ +K +R+W GS + +GHT ++ L +
Sbjct: 31 VVFKGHKRGIWSVEFSPVDQCVVTASGDKTIRIWAISDGSCLKTFEGHTSSVLRALFVTR 210
Query: 272 GRVLFI----RIIRFYDKGQQRCVHSYAVHTDSVWALASTPTFSHVYSGGRD 319
G + +++ + CV +Y H D VWALA + +GG D
Sbjct: 211 GTQIVSCGADGLVKLWTVKTNECVATYDHHEDKVWALAVGRKTEKLATGGGD 366
Score = 37.0 bits (84), Expect = 0.032
Identities = 21/63 (33%), Positives = 34/63 (53%)
Frame = +1
Query: 95 DNTLVSCSSDTTLKTWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVW 154
D +V+ S D T++ W A+S G+C++T HT V + + IV S G G V +W
Sbjct: 85 DQCVVTASGDKTIRIW-AISDGSCLKTFEGHTSSVLRALFVTRGTQIV-SCGADGLVKLW 258
Query: 155 DIE 157
++
Sbjct: 259 TVK 267
>TC220739 weakly similar to UP|PRP4_HUMAN (O43172) U4/U6 small nuclear
ribonucleoprotein Prp4 (U4/U6 snRNP 60 kDa protein) (WD
splicing factor Prp4) (hPrp4), partial (34%)
Length = 1206
Score = 53.1 bits (126), Expect = 4e-07
Identities = 75/346 (21%), Positives = 124/346 (35%), Gaps = 20/346 (5%)
Frame = +1
Query: 47 TISDGSDYLFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAVL--VGDNTLVSCSSD 104
+ S +L T S G K W++ + + F+ H + D V D+ L + S+D
Sbjct: 94 SFSRDGKWLATCSLTGASKLWSMPKIKK--HSIFKGHTERATDVAYSPVHDH-LATASAD 264
Query: 105 TTLKTWNALSAGTCVRTHRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALAPVS 164
T K WN G+ ++T H D + +A + + + +WDIE
Sbjct: 265 RTAKYWNQ---GSLLKTFEGHLDRLARIAFH-PSGKYLGTASFDKTWRLWDIETG----- 417
Query: 165 KCNDAMVDESSNGINGSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAKGHKESVYAL 224
DE + +GH SVY L
Sbjct: 418 -------DEL-------------------------------------LLQEGHSRSVYGL 465
Query: 225 AMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALLLDSTGRVLFI----RII 280
A GS+ S G + + RVWD R+G IL L+GH + + G L
Sbjct: 466 AFHNDGSLAASCGLDSLARVWDLRTGRSILALEGHVKPVLGISFSPNGYHLATGGEDNTC 645
Query: 281 RFYDKGQQRCVHSYAVHTDSV------------WALASTPTFSHVYSGGRDFS--LYLTD 326
R +D +++ ++ H++ + AS + V+S GRDF L+
Sbjct: 646 RIWDLRKKKSFYTIPAHSNLISQVKFEPHEGYFLVTASYDMTAKVWS-GRDFKPVKTLSG 822
Query: 327 LQTRESSLLCTGEHPIRQLALHNDSIWVASTDSSVHRWPAEGCDPQ 372
+ + +S+ G+ SI S D ++ W + D Q
Sbjct: 823 HEAKVTSVDVLGD---------GGSIVTVSHDRTIKLWSSNPTDEQ 933
>TC211497 weakly similar to UP|Q9FNZ2 (Q9FNZ2) Zfwd1 protein (Fragment),
partial (31%)
Length = 840
Score = 50.8 bits (120), Expect = 2e-06
Identities = 32/101 (31%), Positives = 47/101 (45%), Gaps = 2/101 (1%)
Frame = +3
Query: 32 TKHCAGINCLALLTSTISDGSDYLFTGSRDGRLKRWALAEDAATCSATFESHVDWVNDAV 91
T H + CL + G L++GS D +K W + D C+ T H D V ++
Sbjct: 165 TGHTKAVVCLTI-------GCKMLYSGSMDQSIKVWDM--DTLQCTMTLNEHTDIVT-SL 314
Query: 92 LVGDNTLVSCSSDTTLKTWNALSAGT--CVRTHRQHTDYVT 130
+ D L+SCSSD T+K W G+ V TH + V+
Sbjct: 315 ICWDQYLLSCSSDCTIKVWACTEVGSLKVVYTHTEENGVVS 437
Score = 32.7 bits (73), Expect = 0.60
Identities = 27/119 (22%), Positives = 56/119 (46%), Gaps = 7/119 (5%)
Frame = +3
Query: 216 GHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSKILKLKGHTDNIRALL-----LDS 270
GH ++V L + G +L SG ++ ++VWD + + L HTD + +L+ L S
Sbjct: 168 GHTKAVVCLTI--GCKMLYSGSMDQSIKVWDMDTLQCTMTLNEHTDIVTSLICWDQYLLS 341
Query: 271 TGRVLFIRIIRFYDKGQQRCVHSYAVHTD--SVWALASTPTFSHVYSGGRDFSLYLTDL 327
I++ + G + V+++ S++ + ++S RD S+++ +L
Sbjct: 342 CSSDCTIKVWACTEVGSLKVVYTHTEENGVVSLFGMPDAEGKHILFSSCRDNSVHMYEL 518
>TC222379 homologue to UP|Q9HGV7 (Q9HGV7) Gbeta like protein, partial (84%)
Length = 950
Score = 50.1 bits (118), Expect = 4e-06
Identities = 60/252 (23%), Positives = 97/252 (37%), Gaps = 6/252 (2%)
Frame = +1
Query: 4 VASTGNANNSTRPRKEKRLTYVLNDSDDTKHCAGINCLALLTSTISDGSDYLFTGSRDGR 63
++S G S+ K RL + L+ T+ G N L S +D + +GSRD
Sbjct: 58 ISSDGAYALSSSWDKTLRL-WELSTGQTTRRFVGHNNDVLSVSFSADNRQ-IVSGSRDRT 231
Query: 64 LKRWALAEDAATCSATFESHVDWVNDAVLVGDN----TLVSCSSDTTLKTWNALSAGTC- 118
+K W D + T + H +WV+ V N +VS D +K W TC
Sbjct: 232 IKLWNTLGDCKY-TITDKGHTEWVS-CVRFSPNPQNPVIVSAGWDKFVKVWEL---ATCR 396
Query: 119 VRT-HRQHTDYVTCLAAAGKNSNIVASGGLGGEVFVWDIEAALAPVSKCNDAMVDESSNG 177
++T H HT Y+ + + S + ASGG G +WD+ + S +
Sbjct: 397 IQTDHIGHTGYINTVTISPDGS-LCASGGKDGTTMLWDLNESKHLYSLTAGDEIHALVFS 573
Query: 178 INGSANLLPSTNLRTISSSHSISLHTAQTQGYIPIAAKGHKESVYALAMDEGGSILVSGG 237
N +++ + S Y+ + K + +LA G L +G
Sbjct: 574 PNRYWXCAATSSSIIVFDLEKKSKVDELKPEYVEVGKKAREPECISLAWSADGQTLFAGY 753
Query: 238 TEKVVRVWDTRS 249
T+ +R W S
Sbjct: 754 TDNKIRAWGVMS 789
Score = 43.5 bits (101), Expect = 3e-04
Identities = 34/154 (22%), Positives = 64/154 (41%), Gaps = 8/154 (5%)
Frame = +1
Query: 216 GHKESVYALAMDEGGSILVSGGTEKVVRVWDTRSGSK-ILKLKGHTDNIRALLLDSTGRV 274
GH V +++ +VSG ++ +++W+T K + KGHT+ + + +
Sbjct: 154 GHNNDVLSVSFSADNRQIVSGSRDRTIKLWNTLGDCKYTITDKGHTEWVSCVRFSPNPQN 333
Query: 275 LFI------RIIRFYDKGQQRCVHSYAVHTDSVWALASTPTFSHVYSGGRDFSLYLTDLQ 328
I + ++ ++ R + HT + + +P S SGG+D + L DL
Sbjct: 334 PVIVSAGWDKFVKVWELATCRIQTDHIGHTGYINTVTISPDGSLCASGGKDGTTMLWDLN 513
Query: 329 TRESSLLCTGEHPIRQLALHNDSIW-VASTDSSV 361
+ T I L + W A+T SS+
Sbjct: 514 ESKHLYSLTAGDEIHALVFSPNRYWXCAATSSSI 615
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.135 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 34,775,291
Number of Sequences: 63676
Number of extensions: 506209
Number of successful extensions: 2733
Number of sequences better than 10.0: 137
Number of HSP's better than 10.0 without gapping: 2488
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2672
length of query: 744
length of database: 12,639,632
effective HSP length: 104
effective length of query: 640
effective length of database: 6,017,328
effective search space: 3851089920
effective search space used: 3851089920
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC148345.14


