

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148343.5 - phase: 0
(443 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
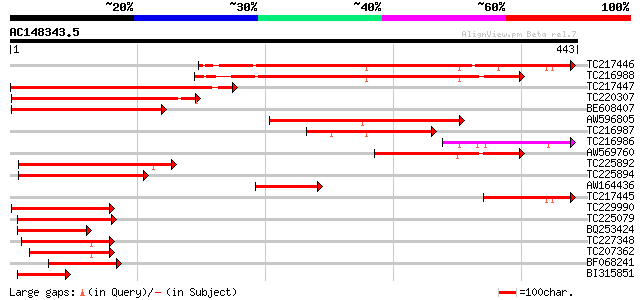
Score E
Sequences producing significant alignments: (bits) Value
TC217446 similar to GB|AAM65362.1|21655301|AY103310 At2g37550/F1... 390 e-109
TC216988 similar to UP|Q84PA3 (Q84PA3) ADP ribosylation GTPase-l... 335 3e-92
TC217447 similar to GB|AAM65362.1|21655301|AY103310 At2g37550/F1... 291 5e-79
TC220307 similar to GB|AAM65362.1|21655301|AY103310 At2g37550/F1... 252 3e-67
BE608407 similar to GP|4519792|dbj Asp1 {Arabidopsis thaliana}, ... 216 2e-56
AW596805 similar to GP|29367415|gb ADP ribosylation GTPase-like ... 211 7e-55
TC216987 similar to UP|Q9FVH2 (Q9FVH2) ARF GAP-like zinc finger-... 163 1e-40
TC216986 127 1e-29
AW569760 weakly similar to GP|29367415|gb| ADP ribosylation GTPa... 125 3e-29
TC225892 similar to UP|ARG3_HUMAN (Q9NP61) ADP-ribosylation fact... 122 3e-28
TC225894 similar to UP|Q9FIQ0 (Q9FIQ0) Zinc finger protein Glo3-... 120 9e-28
AW164436 similar to GP|4519792|dbj| Asp1 {Arabidopsis thaliana},... 102 3e-22
TC217445 weakly similar to GB|AAM65362.1|21655301|AY103310 At2g3... 96 3e-20
TC229990 similar to GB|AAP68261.1|31711810|BT008822 At4g21160 {A... 89 4e-18
TC225079 similar to UP|Q8L8M0 (Q8L8M0) ARF GAP-like zinc finger-... 88 9e-18
BQ253424 81 1e-15
TC227348 similar to UP|Q9SMX5 (Q9SMX5) GCN4-complementing protei... 73 2e-13
TC207362 similar to UP|Q9FIT8 (Q9FIT8) GCN4-complementing protei... 69 3e-12
BF068241 62 4e-10
BI315851 54 1e-07
>TC217446 similar to GB|AAM65362.1|21655301|AY103310 At2g37550/F13M22.5
{Arabidopsis thaliana;} , partial (43%)
Length = 1651
Score = 390 bits (1001), Expect = e-109
Identities = 211/347 (60%), Positives = 247/347 (70%), Gaps = 52/347 (14%)
Frame = +3
Query: 148 GGWDDNWDNDDGDSYGYGS-RGGGDIRRNQSTGDVIGFVGNGVTPSSRSKSTEDIYTRTQ 206
GGWD NWD DDG +GS RGG D+RRNQSTGDV GF G G +RSKST+DIYTR +
Sbjct: 6 GGWD-NWDADDG----FGSARGGSDLRRNQSTGDVRGFGGGGNV--ARSKSTQDIYTRAE 164
Query: 207 LEASAANKEGFFAKKMAENESRPEGLPPSQGGKYVGFGSSPGPAQRISPQNDYLSVVSEG 266
LEASAANKE FFA+K AENESRPEGLPPSQGGKYVGFGS P P QR + QNDY SVVS+G
Sbjct: 165 LEASAANKEEFFARKRAENESRPEGLPPSQGGKYVGFGSGPAPPQRGNQQNDYFSVVSQG 344
Query: 267 IGKLSMVAQSA-------TKEITAKVKDGGYDHKVNETVNIVTQKTSEIGQRTWGIMKGV 319
+GKLS+ A SA TKE++AKVK+GGYD+KVNETVN+V+QKT+EIGQRTWG+MKGV
Sbjct: 345 LGKLSLAAASAANVVQAGTKELSAKVKEGGYDYKVNETVNVVSQKTTEIGQRTWGMMKGV 524
Query: 320 MALASQKVEEFTKDYPDGNSDNWPRNENDRH----DFNQENKGWNSSTSTREGQPSSGGQ 375
MALASQKVEEF +D P+ SDNW RNE+DR+ +FN+EN G NS STREGQ SSGGQ
Sbjct: 525 MALASQKVEEFARDNPNWTSDNWQRNESDRNGFYQEFNRENNGRNS--STREGQHSSGGQ 698
Query: 376 TNTYH----------------------------------SNSWDDWDNQDTRKEVPAKGS 401
TNT+H S+SWDDWD +DTRK+ PAKGS
Sbjct: 699 TNTFHSSSWDDWGHNDFRKEEPAKGSAPQSSGGKFDNYNSSSWDDWDQEDTRKKEPAKGS 878
Query: 402 APHNNDDWAGWDDAKDD--DDEF----DDKSFGNNGTSGSGWTGGGF 442
+ ++D WAGWD+AKDD DD + + K +N SG+ WTGGGF
Sbjct: 879 SARSSDGWAGWDNAKDDGFDDIYGSASNSKGVTHNDKSGAAWTGGGF 1019
>TC216988 similar to UP|Q84PA3 (Q84PA3) ADP ribosylation GTPase-like protein
(Fragment), partial (55%)
Length = 803
Score = 335 bits (858), Expect = 3e-92
Identities = 178/270 (65%), Positives = 204/270 (74%), Gaps = 12/270 (4%)
Frame = +1
Query: 145 SNGGGWDDNWDNDDGDSYGYGSRGGGDIRRNQSTGDVIGFVGNGVTPSSRSKSTEDIYTR 204
+N GGWDD W N+ +++STGD G G P+ RS+STEDIYTR
Sbjct: 7 NNEGGWDD-WGNEASPR-----------SKSKSTGDFRNLNGGGGAPA-RSRSTEDIYTR 147
Query: 205 TQLEASAANKEGFFAKKMAENESRPEGLPPSQGGKYVGFGSSPGPAQRISP-QNDYLSVV 263
TQLEASAA KE FFA+K+AENESRPEGLPPSQGGKYVGFGSSP P QR + QNDY SVV
Sbjct: 148 TQLEASAAKKEDFFARKIAENESRPEGLPPSQGGKYVGFGSSPQPPQRRNEAQNDYFSVV 327
Query: 264 SEGIGKLSMVAQSA-------TKEITAKVKDGGYDHKVNETVNIVTQKTSEIGQRTWGIM 316
S+GIGKLS+VA SA TKE T+KVK+GGYDHKVNETVN+VTQKTSEIGQRTWGIM
Sbjct: 328 SQGIGKLSLVAASAANVVQAGTKEFTSKVKEGGYDHKVNETVNVVTQKTSEIGQRTWGIM 507
Query: 317 KGVMALASQKVEEFTKDYPDGNSDNWPRNENDRH----DFNQENKGWNSSTSTREGQPSS 372
KGVMA+ASQK+EE+TKD P+ +DNW RNENDR+ DFNQENKGWNSS + SS
Sbjct: 508 KGVMAMASQKIEEYTKDDPNWKNDNWQRNENDRNGYYQDFNQENKGWNSSIGREQ---SS 678
Query: 373 GGQTNTYHSNSWDDWDNQDTRKEVPAKGSA 402
GQ T S+SWDDWD+ D+RKEVPAKGS+
Sbjct: 679 SGQFKTQSSSSWDDWDHNDSRKEVPAKGSS 768
>TC217447 similar to GB|AAM65362.1|21655301|AY103310 At2g37550/F13M22.5
{Arabidopsis thaliana;} , partial (35%)
Length = 563
Score = 291 bits (744), Expect = 5e-79
Identities = 140/179 (78%), Positives = 156/179 (86%), Gaps = 1/179 (0%)
Frame = +3
Query: 1 MAASRRLRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVT 60
MAASRRLR+LQS+P+NKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVT
Sbjct: 39 MAASRRLRDLQSQPANKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVT 218
Query: 61 MDSWSDLQIKKMEAGGNRNLNTFLSQYGISKETDIITKYNSNAASIYRDRIQAIAEGRSW 120
MDSWS++QIKKMEAGGN LN FL+QYGI KETDI+ KYNSNAA++YRDRIQA+A+GR W
Sbjct: 219 MDSWSEIQIKKMEAGGNDKLNAFLTQYGIPKETDIVAKYNSNAAAVYRDRIQALADGRPW 398
Query: 121 RDPPVVKENASTRAGKGKPPLAAASNGGGWDDNWDNDDGDSYGYGS-RGGGDIRRNQST 178
RDPPVVKE + A KGKPPL+A +N G DNWD DD G+GS RGG D+RRNQST
Sbjct: 399 RDPPVVKEAVRSSASKGKPPLSAGNNNNGGWDNWDPDD----GFGSARGGADLRRNQST 563
>TC220307 similar to GB|AAM65362.1|21655301|AY103310 At2g37550/F13M22.5
{Arabidopsis thaliana;} , partial (28%)
Length = 602
Score = 252 bits (643), Expect = 3e-67
Identities = 124/151 (82%), Positives = 133/151 (87%), Gaps = 3/151 (1%)
Frame = +2
Query: 2 AASRRLRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTM 61
AASRRLR+LQSE NKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTM
Sbjct: 152 AASRRLRDLQSEAGNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTM 331
Query: 62 DSWSDLQIKKMEAGGNRNLNTFLSQYGISKETDIITKYNSNAASIYRDRIQAIAEGRSWR 121
DSWSD+QIKKMEAGGN LN FL+QY I KETDI+TKYN+NAAS+YR+RIQAIAEGR WR
Sbjct: 332 DSWSDIQIKKMEAGGNDKLNAFLAQYSIPKETDIVTKYNTNAASVYRNRIQAIAEGRPWR 511
Query: 122 DPPVVKENASTRAGKGKPPLAAA---SNGGG 149
DPPV+KEN S AGKGKPPL +N GG
Sbjct: 512 DPPVLKENLSA-AGKGKPPLTQTRRNNNEGG 601
>BE608407 similar to GP|4519792|dbj Asp1 {Arabidopsis thaliana}, partial
(26%)
Length = 421
Score = 216 bits (550), Expect = 2e-56
Identities = 101/121 (83%), Positives = 111/121 (91%)
Frame = +2
Query: 2 AASRRLRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTM 61
AASRRLR+LQS+P+NKICVDCSQKNPQWASVSYGVFMCLECSGKHRGL VHI FVRSVTM
Sbjct: 2 AASRRLRDLQSQPANKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLCVHICFVRSVTM 181
Query: 62 DSWSDLQIKKMEAGGNRNLNTFLSQYGISKETDIITKYNSNAASIYRDRIQAIAEGRSWR 121
DSWS++QIKKMEAGGN LN FL QYGI KETDI+ KY+SNAAS+YRDRI A+A+GR WR
Sbjct: 182 DSWSEIQIKKMEAGGNDKLNAFLLQYGIPKETDIVVKYSSNAASVYRDRIHALADGRPWR 361
Query: 122 D 122
D
Sbjct: 362 D 364
>AW596805 similar to GP|29367415|gb ADP ribosylation GTPase-like protein
{Oryza sativa (japonica cultivar-group)}, partial (33%)
Length = 472
Score = 211 bits (536), Expect = 7e-55
Identities = 108/157 (68%), Positives = 125/157 (78%), Gaps = 5/157 (3%)
Frame = +2
Query: 204 RTQLEASAANKEGFFAKKMAENESRPEGLPPSQGGKYVGFGSSPGPAQR-ISPQNDYLSV 262
R +LEA AANKE FFA+K AENESRPEGLPP QGGKYVGF S P P Q ++ QNDYLSV
Sbjct: 2 RAELEACAANKEDFFARKRAENESRPEGLPPPQGGKYVGF*SGPAPNQTGVNQQNDYLSV 181
Query: 263 VSEGIGKLSMVA----QSATKEITAKVKDGGYDHKVNETVNIVTQKTSEIGQRTWGIMKG 318
VS G GKLS+ A Q+ TKE +AKVK+GGYD+KVNETVN+V+ KTSEIGQRTWGIMKG
Sbjct: 182 VSHGFGKLSLAAANVVQAGTKEFSAKVKEGGYDYKVNETVNVVSHKTSEIGQRTWGIMKG 361
Query: 319 VMALASQKVEEFTKDYPDGNSDNWPRNENDRHDFNQE 355
VMALASQ VEEF++D P+ SD+W R E+DR+ E
Sbjct: 362 VMALASQNVEEFSRDNPNWMSDDWRRIESDRNGLYHE 472
>TC216987 similar to UP|Q9FVH2 (Q9FVH2) ARF GAP-like zinc finger-containing
protein ZIGA2 (Fragment), partial (23%)
Length = 337
Score = 163 bits (413), Expect = 1e-40
Identities = 84/110 (76%), Positives = 92/110 (83%), Gaps = 9/110 (8%)
Frame = +3
Query: 233 PPSQGGKYVGFGSSPGPA--QRISPQNDYLSVVSEGIGKLSMVAQSA-------TKEITA 283
PPSQGGKYVGFGSSP P +R QNDY SVVS+GIGKLS VA SA TKE T+
Sbjct: 6 PPSQGGKYVGFGSSPTPPPQRRNDAQNDYFSVVSQGIGKLSFVAASAANVVQVGTKEFTS 185
Query: 284 KVKDGGYDHKVNETVNIVTQKTSEIGQRTWGIMKGVMALASQKVEEFTKD 333
KVK+GGYDHKVNETVN+VTQKTSEIGQRTWGIMKGVMA+ASQK+EE+TKD
Sbjct: 186 KVKEGGYDHKVNETVNVVTQKTSEIGQRTWGIMKGVMAMASQKIEEYTKD 335
>TC216986
Length = 914
Score = 127 bits (318), Expect = 1e-29
Identities = 67/143 (46%), Positives = 84/143 (57%), Gaps = 39/143 (27%)
Frame = +1
Query: 339 SDNWPRNENDRH----DFNQENKGWNSSTS------------------------TREGQP 370
+DNW RNEND + DFN+E KGWNSST +R+ +P
Sbjct: 1 NDNWQRNENDTNGYYQDFNREKKGWNSSTGREQSSSGEYKTESSSSWDDWDHKDSRKEEP 180
Query: 371 ------SSGGQTNTYHSNSWDDWDNQDTRKEVPAKGSAPHNNDDWAGWDDAKDDD----- 419
SSGGQ +TY+S+SWDDW+++D+RKE PAKGSAPHNNDDWAGWD+AKDD
Sbjct: 181 AKGSTHSSGGQFSTYNSSSWDDWEHKDSRKEEPAKGSAPHNNDDWAGWDNAKDDGFDNIY 360
Query: 420 DEFDDKSFGNNGTSGSGWTGGGF 442
+ + K G+ G S WT GF
Sbjct: 361 EGANKKGVGHKGKSDGTWTEEGF 429
>AW569760 weakly similar to GP|29367415|gb| ADP ribosylation GTPase-like
protein {Oryza sativa (japonica cultivar-group)},
partial (22%)
Length = 358
Score = 125 bits (315), Expect = 3e-29
Identities = 66/121 (54%), Positives = 81/121 (66%), Gaps = 4/121 (3%)
Frame = +2
Query: 286 KDGGYDHKVNETVNIVTQKTSEIGQRTWGIMKGVMALASQKVEEFTKDYPDGNSDNWPRN 345
K+GGY+H VNETVN+ TQKTS+I QRTWGIM GV+A+A Q +EE+TK P+ +D W RN
Sbjct: 2 KEGGYNHMVNETVNVATQKTSKIVQRTWGIMIGVIAIAPQNIEEYTKHDPNCKNDYWQRN 181
Query: 346 END----RHDFNQENKGWNSSTSTREGQPSSGGQTNTYHSNSWDDWDNQDTRKEVPAKGS 401
END DFN + RE SS GQ T S+SWDDWD D+ KEVP+KGS
Sbjct: 182 ENDINGYYQDFNLIEQLAGILLLVRE--QSSSGQFKTQISSSWDDWDRNDSGKEVPSKGS 355
Query: 402 A 402
+
Sbjct: 356 S 358
>TC225892 similar to UP|ARG3_HUMAN (Q9NP61) ADP-ribosylation factor
GTPase-activating protein 3 (ARF GAP 3), partial (12%)
Length = 1727
Score = 122 bits (306), Expect = 3e-28
Identities = 58/128 (45%), Positives = 81/128 (62%), Gaps = 5/128 (3%)
Frame = +2
Query: 8 RELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSWSDL 67
R+L+++ NK+C DC+ KNP WASV+YG+F+C++CS HR LGVHISFVRS +DSWS
Sbjct: 248 RKLKAKSENKMCFDCNAKNPTWASVTYGIFLCIDCSAVHRSLGVHISFVRSTNLDSWSPE 427
Query: 68 QIKKMEAGGNRNLNTFLSQYGISKETDIITKYNSNAASIYRDRI-----QAIAEGRSWRD 122
Q+K M GGN + F Q+G + I KY S AA +YR + +++AE
Sbjct: 428 QLKTMSFGGNNRAHGFFKQHGWTDGGKIEAKYTSRAADLYRQILSKEVAKSMAEDGGLPS 607
Query: 123 PPVVKENA 130
PV ++A
Sbjct: 608 SPVASQSA 631
>TC225894 similar to UP|Q9FIQ0 (Q9FIQ0) Zinc finger protein Glo3-like,
partial (32%)
Length = 612
Score = 120 bits (302), Expect = 9e-28
Identities = 53/101 (52%), Positives = 69/101 (67%)
Frame = +1
Query: 8 RELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSWSDL 67
R+L+++ NK+C DC+ KNP WASV+YG+F+C++CS HR LGVHISFVRS +DSWS
Sbjct: 247 RKLKAKSENKMCFDCNAKNPTWASVTYGIFLCIDCSAVHRSLGVHISFVRSTNLDSWSPE 426
Query: 68 QIKKMEAGGNRNLNTFLSQYGISKETDIITKYNSNAASIYR 108
Q+K M GGN F Q+G + I KY S AA +YR
Sbjct: 427 QLKTMSFGGNNRAQVFFKQHGWNDGGKIEAKYTSRAADLYR 549
>AW164436 similar to GP|4519792|dbj| Asp1 {Arabidopsis thaliana}, partial
(11%)
Length = 163
Score = 102 bits (254), Expect = 3e-22
Identities = 48/52 (92%), Positives = 52/52 (99%)
Frame = +3
Query: 193 SRSKSTEDIYTRTQLEASAANKEGFFAKKMAENESRPEGLPPSQGGKYVGFG 244
+RS+STEDIYTR+QLEASAANKEGFFA+KMAENESRPEGLPPSQGGKYVGFG
Sbjct: 6 ARSRSTEDIYTRSQLEASAANKEGFFARKMAENESRPEGLPPSQGGKYVGFG 161
>TC217445 weakly similar to GB|AAM65362.1|21655301|AY103310
At2g37550/F13M22.5 {Arabidopsis thaliana;} , partial
(5%)
Length = 645
Score = 95.9 bits (237), Expect = 3e-20
Identities = 45/79 (56%), Positives = 58/79 (72%), Gaps = 7/79 (8%)
Frame = +2
Query: 371 SSGGQ-TNTYHSNSWDDWDNQDTRKEVPAKGSAPHNNDDWAGWDDAKDD--DDEF----D 423
SSGG+ N Y+S+SWDDWD +DTRK+ PAKGSA H++D W GWD+AKDD DD + +
Sbjct: 2 SSGGKFDNNYNSSSWDDWDQEDTRKKEPAKGSAAHSSDGWTGWDNAKDDGFDDIYGSASN 181
Query: 424 DKSFGNNGTSGSGWTGGGF 442
+K +N SG+ WTGGGF
Sbjct: 182 NKGGAHNDKSGAAWTGGGF 238
>TC229990 similar to GB|AAP68261.1|31711810|BT008822 At4g21160 {Arabidopsis
thaliana;} , partial (22%)
Length = 444
Score = 89.0 bits (219), Expect = 4e-18
Identities = 39/82 (47%), Positives = 58/82 (70%), Gaps = 1/82 (1%)
Frame = +3
Query: 2 AASRRLRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTM 61
++ R+L++L + N++C DC+ +P+WAS + GVF+CL+C G HR LG HIS V SVT+
Sbjct: 198 SSRRKLKDLLLQSDNRLCADCNAPDPKWASANIGVFICLKCCGVHRSLGTHISKVLSVTL 377
Query: 62 DSWSDLQIKKM-EAGGNRNLNT 82
D WS+ +I M E GGN + N+
Sbjct: 378 DDWSEDEIDAMIEVGGNASANS 443
>TC225079 similar to UP|Q8L8M0 (Q8L8M0) ARF GAP-like zinc finger-containing
protein ZIGA3, partial (48%)
Length = 1845
Score = 87.8 bits (216), Expect = 9e-18
Identities = 38/77 (49%), Positives = 51/77 (65%)
Frame = +3
Query: 7 LRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSWSD 66
L L P N+ C DC K P+WASV+ G+F+C++CSG HR LGVHIS VRS T+D+W
Sbjct: 189 LEGLLKLPENRECADCKAKGPRWASVNLGIFICMQCSGIHRSLGVHISKVRSATLDTWLP 368
Query: 67 LQIKKMEAGGNRNLNTF 83
Q+ +++ GN N F
Sbjct: 369 EQVAFIQSMGNEKANCF 419
>BQ253424
Length = 277
Score = 80.9 bits (198), Expect = 1e-15
Identities = 34/58 (58%), Positives = 42/58 (71%)
Frame = +2
Query: 7 LRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSW 64
L L P NK C DC K P+WASV+ G+F+C++CSG HR LGVHIS VRS T+D+W
Sbjct: 104 LEGLLKLPENKECADCKAKGPRWASVNLGIFICMQCSGIHRSLGVHISKVRSATLDTW 277
>TC227348 similar to UP|Q9SMX5 (Q9SMX5) GCN4-complementing protein (GCP1),
partial (29%)
Length = 1197
Score = 73.2 bits (178), Expect = 2e-13
Identities = 32/75 (42%), Positives = 47/75 (62%), Gaps = 2/75 (2%)
Frame = +3
Query: 10 LQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMD--SWSDL 67
L+ P N C +CS P WAS++ G+ +C+ECSG HR LGVH+S VRS+T+D W +
Sbjct: 576 LRGIPGNDKCAECSAPEPDWASLNLGILLCIECSGVHRNLGVHVSKVRSITLDVRVWENT 755
Query: 68 QIKKMEAGGNRNLNT 82
++ + GN N+
Sbjct: 756 VLELFDNLGNAYCNS 800
>TC207362 similar to UP|Q9FIT8 (Q9FIT8) GCN4-complementing protein homolog,
partial (51%)
Length = 1959
Score = 69.3 bits (168), Expect = 3e-12
Identities = 33/69 (47%), Positives = 42/69 (60%), Gaps = 2/69 (2%)
Frame = +1
Query: 16 NKICVDCSQKNPQWASVSYGVFMCLECSGKHRGLGVHISFVRSVTMD--SWSDLQIKKME 73
N C DC P WAS++ GV +C+ECSG HR LGVHIS VRS+T+D W I +
Sbjct: 793 NDKCADCGAPEPDWASLNLGVLVCIECSGVHRNLGVHISKVRSLTLDVKVWEPSVISLFQ 972
Query: 74 AGGNRNLNT 82
+ GN N+
Sbjct: 973 SLGNTFANS 999
>BF068241
Length = 387
Score = 62.4 bits (150), Expect = 4e-10
Identities = 30/58 (51%), Positives = 38/58 (64%), Gaps = 1/58 (1%)
Frame = +2
Query: 31 SVSYGVFMCLECSGKHRGLGVHISFVRSVTMDSWSDLQI-KKMEAGGNRNLNTFLSQY 87
S + GVF+CL+C G HR LG HIS V SVT+D WS+ +I ME GGN + N+ Y
Sbjct: 188 SANIGVFICLKCCGVHRSLGTHISKVLSVTLDDWSEDEIDAMMEVGGNASANSIYEAY 361
>BI315851
Length = 385
Score = 53.9 bits (128), Expect = 1e-07
Identities = 21/41 (51%), Positives = 28/41 (68%)
Frame = +1
Query: 7 LRELQSEPSNKICVDCSQKNPQWASVSYGVFMCLECSGKHR 47
L L P N+ C DC K P+WASV+ G+F+C++CSG HR
Sbjct: 262 LEGLLKLPENRECADCRNKAPRWASVNLGIFICMQCSGIHR 384
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.309 0.129 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,950,525
Number of Sequences: 63676
Number of extensions: 268566
Number of successful extensions: 1726
Number of sequences better than 10.0: 85
Number of HSP's better than 10.0 without gapping: 1506
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1657
length of query: 443
length of database: 12,639,632
effective HSP length: 100
effective length of query: 343
effective length of database: 6,272,032
effective search space: 2151306976
effective search space used: 2151306976
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC148343.5


