

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148342.7 - phase: 0
(780 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
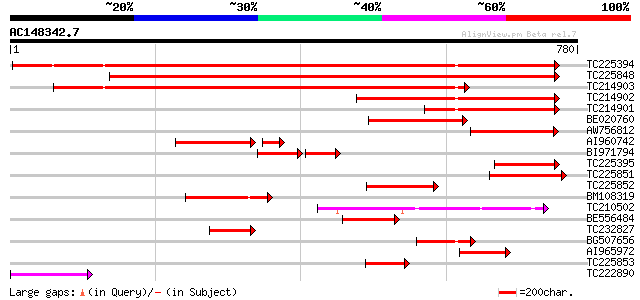
Score E
Sequences producing significant alignments: (bits) Value
TC225394 UP|SUSY_SOYBN (P13708) Sucrose synthase (Sucrose-UDP g... 866 0.0
TC225848 homologue to UP|SUS2_PEA (O24301) Sucrose synthase 2 (... 788 0.0
TC214903 similar to UP|Q9T0M9 (Q9T0M9) Sucrose synthase , parti... 651 0.0
TC214902 homologue to UP|Q9AVR8 (Q9AVR8) Sucrose synthase isofor... 392 e-109
TC214901 homologue to UP|SUSY_SOYBN (P13708) Sucrose synthase (... 258 5e-69
BE020760 226 4e-59
AW756812 161 9e-40
AI960742 homologue to GP|4098128|gb| sucrose synthase {Gossypium... 137 2e-38
BI971794 homologue to SP|P13708|SUSY Sucrose synthase (EC 2.4.1.... 86 4e-35
TC225395 homologue to UP|SUSY_SOYBN (P13708) Sucrose synthase (... 141 1e-33
TC225851 similar to UP|SUS2_PEA (O24301) Sucrose synthase 2 (Su... 134 2e-31
TC225852 similar to UP|Q9SLY2 (Q9SLY2) Sucrose synthase, partial... 133 3e-31
BM108319 homologue to SP|P13708|SUSY_ Sucrose synthase (EC 2.4.1... 113 3e-25
TC210502 homologue to UP|Q9AXK3 (Q9AXK3) Sucrose-phosphate synth... 112 6e-25
BE556484 similar to SP|P13708|SUSY_ Sucrose synthase (EC 2.4.1.1... 101 1e-21
TC232827 similar to UP|Q94G60 (Q94G60) Sucrose synthase , parti... 95 1e-19
BG507656 similar to SP|P13708|SUSY_ Sucrose synthase (EC 2.4.1.1... 90 3e-18
AI965972 similar to SP|P13708|SUSY_ Sucrose synthase (EC 2.4.1.1... 89 6e-18
TC225853 homologue to UP|Q9SLY2 (Q9SLY2) Sucrose synthase, parti... 88 1e-17
TC222890 similar to UP|SUS2_PEA (O24301) Sucrose synthase 2 (Su... 88 2e-17
>TC225394 UP|SUSY_SOYBN (P13708) Sucrose synthase (Sucrose-UDP
glucosyltransferase) (Nodulin-100) , complete
Length = 3328
Score = 866 bits (2238), Expect = 0.0
Identities = 427/754 (56%), Positives = 558/754 (73%), Gaps = 1/754 (0%)
Frame = +1
Query: 4 THALKRTNSIADNMPDALRKSRYHMKKCFAKYLEKGRRIMKLHELMEEVERTIDDINERN 63
T L R +S+ + + + L +R + ++ KG+ I++ H+++ E E ++ R
Sbjct: 73 TDRLTRVHSLRERLDETLTANRNEILALLSRIEAKGKGILQHHQVIAEFEEIPEE--NRQ 246
Query: 64 YILEGNLGFILSSTQEAVVDPPYVAFAIRPNPGVWEYVRVNSEDLSVEPITPTDYLKFKE 123
+ +G G +L STQEA+V PP+VA A+RP PGVWEY+RVN L VE + P +YL FKE
Sbjct: 247 KLTDGAFGEVLRSTQEAIVLPPWVALAVRPRPGVWEYLRVNVHALVVEELQPAEYLHFKE 426
Query: 124 RVYDQKWANDENAFEADFGAFDIGIPKLTLSSSIGNGLHFVSKFLTSRTTGKLAKAQTIV 183
+ D +N E DF F+ P+ TL+ SIGNG+ F+++ L+++ ++
Sbjct: 427 ELVDGS-SNGNFVLELDFEPFNAAFPRPTLNKSIGNGVQFLNRHLSAKLFHDKESLHPLL 603
Query: 184 DYLLKLNHHGESLMINDTLSSAAKLQMALIVADVFLSAIPKDTSYQKFELRLKEWGFEKG 243
++L + G++LM+ND + + LQ L A+ +L +P +T Y +FE + +E G E+G
Sbjct: 604 EFLRLHSVKGKTLMLNDRIQNPDALQHVLRKAEEYLGTVPPETPYSEFEHKFQEIGLERG 783
Query: 244 WGDNAGRVKETMRTLSEVLQAPDPVNLEIFFSRIPTIFKVVIFSVHGYFGQADVLGLPDT 303
WGDNA RV E+++ L ++L+APDP LE F RIP +F VVI S HGYF Q +VLG PDT
Sbjct: 784 WGDNAERVLESIQLLLDLLEAPDPCTLETFLGRIPMVFNVVILSPHGYFAQDNVLGYPDT 963
Query: 304 GGQVVYILDQVKALEEELILRIKQQGLNYKPQILVVTRLIPDARGTKCHQEFEPINDTKH 363
GGQVVYILDQV+ALE E++ RIKQQGL+ P+IL++TRL+PDA GT C Q E + T+H
Sbjct: 964 GGQVVYILDQVRALENEMLHRIKQQGLDIVPRILIITRLLPDAIGTTCGQRLEKVFGTEH 1143
Query: 364 SHILRVPFHTEKGILPQWVSRFDIYPYLERFTQDATTKILDLMEGKPDLVIGNYTDGNLV 423
SHILRVPF TEKGI+ +W+SRF+++PYLE +T+D ++ ++GKPDL++GNY+DGN+V
Sbjct: 1144SHILRVPFRTEKGIVRKWISRFEVWPYLETYTEDVAHELAKELQGKPDLIVGNYSDGNIV 1323
Query: 424 ASLMARKLGITQATIAHALEKTKYEDSDVKWKELDPKYHFSCQFMADTVAMNSSDFIITS 483
ASL+A KLG+TQ TIAHALEKTKY +SD+ WK+L+ +YHFSCQF AD AMN +DFIITS
Sbjct: 1324ASLLAHKLGVTQCTIAHALEKTKYPESDIYWKKLEERYHFSCQFTADLFAMNHTDFIITS 1503
Query: 484 TYQEIAGSKDRPGQYESHAAFTLPGLCRVVSGINVFDPKFNIAAPGADQSIYFPYTEKDQ 543
T+QEIAGSKD GQYESH AFTLPGL RVV GI+VFDPKFNI +PGADQ+IYFP TE +
Sbjct: 1504TFQEIAGSKDTVGQYESHTAFTLPGLYRVVHGIDVFDPKFNIVSPGADQTIYFPPTETSR 1683
Query: 544 RHSQFHPAIEDLLFNKVDNNEHIGYLADKRKPIIFSMARLDVVKNLSGLVEWYGKNKRLR 603
R + FHP IE+LL++ V+N EHI L D+ KPIIF+MARLD VKN++GLVEWYGKN +LR
Sbjct: 1684RLTSFHPEIEELLYSSVENEEHICVLKDRSKPIIFTMARLDRVKNITGLVEWYGKNAKLR 1863
Query: 604 NLVNLVIVGGFFDPSK-SKDREEMAEIKKMHDLIEKYQLKGQFRWIAAQTDRYRNGELYR 662
LVNLV+V G D K SKD EE AE+KKM+ LIE Y+L GQFRWI++Q +R RNGELYR
Sbjct: 1864ELVNLVVVAG--DRRKESKDLEEKAEMKKMYGLIETYKLNGQFRWISSQMNRVRNGELYR 2037
Query: 663 CIADTKGAFVQPALYEAFGLTVIEAMNCGLPTFATNQGGPAEIIVDGVSGFHIDPLNGDE 722
I DTKGAFVQPA+YEAFGLTV+EAM CGLPTFAT GGPAEIIV G SGFHIDP +GD
Sbjct: 2038VICDTKGAFVQPAIYEAFGLTVVEAMTCGLPTFATCNGGPAEIIVHGKSGFHIDPYHGDR 2217
Query: 723 SSNKISDFFEKCKVDPSYWNVISMAGLQRINEWY 756
+++ + DFFEKCK+DP++W+ IS AGLQRI E Y
Sbjct: 2218AADLLVDFFEKCKLDPTHWDKISKAGLQRIEEKY 2319
>TC225848 homologue to UP|SUS2_PEA (O24301) Sucrose synthase 2 (Sucrose-UDP
glucosyltransferase 2) , partial (83%)
Length = 2487
Score = 788 bits (2035), Expect = 0.0
Identities = 384/619 (62%), Positives = 477/619 (77%)
Frame = +2
Query: 138 EADFGAFDIGIPKLTLSSSIGNGLHFVSKFLTSRTTGKLAKAQTIVDYLLKLNHHGESLM 197
E DF F+ P+ T S+SIGNG+ F+++ L+S Q ++D+L + G +LM
Sbjct: 20 ELDFEPFNATFPRPTRSASIGNGVQFLNRHLSSIMFRNKDSLQPLLDFLRAHKYKGHALM 199
Query: 198 INDTLSSAAKLQMALIVADVFLSAIPKDTSYQKFELRLKEWGFEKGWGDNAGRVKETMRT 257
+ND + + AKLQ AL A+ +LS + DT Y +FE L+ GFE+GWGD A RV E M
Sbjct: 200 LNDRIQTIAKLQSALAKAEDYLSKLAHDTLYSEFEYVLQGMGFERGWGDTAERVLEMMHL 379
Query: 258 LSEVLQAPDPVNLEIFFSRIPTIFKVVIFSVHGYFGQADVLGLPDTGGQVVYILDQVKAL 317
L ++LQAPDP LE F R+P +F V I S HGYFGQA+VLGLPDTGGQVVYILDQV+AL
Sbjct: 380 LLDILQAPDPSTLETFLGRVPMVFNVAILSPHGYFGQANVLGLPDTGGQVVYILDQVRAL 559
Query: 318 EEELILRIKQQGLNYKPQILVVTRLIPDARGTKCHQEFEPINDTKHSHILRVPFHTEKGI 377
E E++LRIK+QGL++ P+IL+VTRLIPDA+GT C+Q E ++ T H+HILRVPF +E G
Sbjct: 560 ENEMLLRIKKQGLDFTPRILIVTRLIPDAKGTTCNQRLERVSGTDHTHILRVPFRSESGT 739
Query: 378 LPQWVSRFDIYPYLERFTQDATTKILDLMEGKPDLVIGNYTDGNLVASLMARKLGITQAT 437
L +W+SRFD++PYLE + +D ++I ++G PD +IGNY+DGNLVASL+A K+G+TQ T
Sbjct: 740 LRKWISRFDVWPYLETYAEDVASEIAAELQGYPDFIIGNYSDGNLVASLLAYKMGVTQCT 919
Query: 438 IAHALEKTKYEDSDVKWKELDPKYHFSCQFMADTVAMNSSDFIITSTYQEIAGSKDRPGQ 497
IAHALEKTKY DSD+ WK+ + KYHFSCQF AD +AMN++DFIITSTYQEIAG+K+ GQ
Sbjct: 920 IAHALEKTKYPDSDLYWKKFEDKYHFSCQFTADLIAMNNADFIITSTYQEIAGTKNTVGQ 1099
Query: 498 YESHAAFTLPGLCRVVSGINVFDPKFNIAAPGADQSIYFPYTEKDQRHSQFHPAIEDLLF 557
YESHA FTLPGL RVV GI+VFDPKFNI +PGAD SIYFPY+EK R + H +IE LLF
Sbjct: 1100YESHAGFTLPGLYRVVHGIDVFDPKFNIVSPGADMSIYFPYSEKQNRLTALHGSIEQLLF 1279
Query: 558 NKVDNNEHIGYLADKRKPIIFSMARLDVVKNLSGLVEWYGKNKRLRNLVNLVIVGGFFDP 617
+E+IG L DK KPIIFSMARLD VKN++GLVE +GKN +LR LVNLV+V G+ D
Sbjct: 1280APEQTDEYIGLLKDKSKPIIFSMARLDRVKNITGLVESFGKNSKLRELVNLVVVAGYIDV 1459
Query: 618 SKSKDREEMAEIKKMHDLIEKYQLKGQFRWIAAQTDRYRNGELYRCIADTKGAFVQPALY 677
KS DREE+AEI+KMH+L++KY L G FRWIAAQT+R RNGELYR IADT+GAFVQPA Y
Sbjct: 1460KKSSDREEIAEIEKMHELMKKYNLNGDFRWIAAQTNRARNGELYRYIADTQGAFVQPAFY 1639
Query: 678 EAFGLTVIEAMNCGLPTFATNQGGPAEIIVDGVSGFHIDPLNGDESSNKISDFFEKCKVD 737
EAFGLTV+EAMNCGLPTFAT GGPAEII G+SGFHIDP + D++S + +FF+K K D
Sbjct: 1640EAFGLTVVEAMNCGLPTFATCHGGPAEIIEHGISGFHIDPYHPDQASQLLVEFFQKSKED 1819
Query: 738 PSYWNVISMAGLQRINEWY 756
PS+W IS GLQRI E Y
Sbjct: 1820PSHWKKISDGGLQRIYERY 1876
>TC214903 similar to UP|Q9T0M9 (Q9T0M9) Sucrose synthase , partial (78%)
Length = 2572
Score = 651 bits (1679), Expect = 0.0
Identities = 318/573 (55%), Positives = 423/573 (73%), Gaps = 1/573 (0%)
Frame = +1
Query: 61 ERNYILEGNLGFILSSTQEAVVDPPYVAFAIRPNPGVWEYVRVNSEDLSVEPITPTDYLK 120
++ + +G G +L STQEA+V PP+VA A+RP PGVWEY+RVN L V+ + P +YL+
Sbjct: 862 QKENLQDGVFGEVLRSTQEAIVLPPFVALAVRPRPGVWEYLRVNVHMLVVDELRPAEYLR 1041
Query: 121 FKERVYDQKWANDENAFEADFGAFDIGIPKLTLSSSIGNGLHFVSKFLTSRTTGKLAKAQ 180
FKE + + +N E DF F+ P+ TL+ SIGNG+ F+++ L+++ Q
Sbjct: 1042 FKEELVEGS-SNGNFVLELDFEPFNASFPRPTLNKSIGNGVEFLNRHLSAKLFHDKESMQ 1218
Query: 181 TIVDYLLKLNHHGESLMINDTLSSAAKLQMALIVADVFLSAIPKDTSYQKFELRLKEWGF 240
++++L ++ G+++M+ND + S LQ L A+ +L ++ +T Y +FE R +E G
Sbjct: 1219 PLLEFLRLHSYKGKTMMLNDKVQSLDSLQHVLRKAEEYLISVAPETPYSEFENRFREIGL 1398
Query: 241 EKGWGDNAGRVKETMRTLSEVLQAPDPVNLEIFFSRIPTIFKVVIFSVHGYFGQADVLGL 300
E+GWGD A RV E ++ L ++L+APDP LE F R+P +F VVI S HGYF Q +VLG
Sbjct: 1399 ERGWGDTAERVLEMIQLLLDLLEAPDPCTLETFLGRVPMVFNVVILSPHGYFAQDNVLGY 1578
Query: 301 PDTGGQVVYILDQVKALEEELILRIKQQGLNYKPQILVVTRLIPDARGTKCHQEFEPIND 360
PDTGGQVVYILDQV+ALE E++ RIK+QGL+ P+IL++TRL+PDA GT C Q E + D
Sbjct: 1579 PDTGGQVVYILDQVRALENEMLNRIKKQGLDITPRILIITRLLPDAVGTTCGQRLERVYD 1758
Query: 361 TKHSHILRVPFHTEKGILPQWVSRFDIYPYLERFTQDATTKILDLMEGKPDLVIGNYTDG 420
T++ ILRVPF TEKGI+ +W+SRF+++PYLE +T+D ++ ++ KPDL++GNY+DG
Sbjct: 1759 TEYCDILRVPFRTEKGIVRKWISRFEVWPYLETYTEDVALELAKELQAKPDLIVGNYSDG 1938
Query: 421 NLVASLMARKLGITQATIAHALEKTKYEDSDVKWKELDPKYHFSCQFMADTVAMNSSDFI 480
N+VASL+A KLG+TQ TIAHALEKTKY +SD+ WK+ + KYHFSCQF AD AMN +DFI
Sbjct: 1939 NIVASLLAHKLGVTQCTIAHALEKTKYPESDIYWKKFEEKYHFSCQFTADLFAMNHTDFI 2118
Query: 481 ITSTYQEIAGSKDRPGQYESHAAFTLPGLCRVVSGINVFDPKFNIAAPGADQSIYFPYTE 540
ITST+QEIAGSKD GQYESH AFTLPGL RVV GI+ FDPKFNI +PGAD IYFPYTE
Sbjct: 2119 ITSTFQEIAGSKDTVGQYESHTAFTLPGLYRVVHGIDPFDPKFNIVSPGADMGIYFPYTE 2298
Query: 541 KDQRHSQFHPAIEDLLFNKVDNNEHIGYLADKRKPIIFSMARLDVVKNLSGLVEWYGKNK 600
++R ++FH IE+LL++ V+N EHI L D+ KPIIF+MARLD VKN++GLVEWYGKN
Sbjct: 2299 TERRLTEFHSDIEELLYSSVENEEHICVLKDRNKPIIFTMARLDRVKNITGLVEWYGKNA 2478
Query: 601 RLRNLVNLVIVGGFFDPSK-SKDREEMAEIKKM 632
RLR LVNLV+V G D K SKD EE AE+KKM
Sbjct: 2479 RLRELVNLVVVAG--DRRKESKDLEEKAEMKKM 2571
>TC214902 homologue to UP|Q9AVR8 (Q9AVR8) Sucrose synthase isoform 3 ,
partial (41%)
Length = 1430
Score = 392 bits (1007), Expect = e-109
Identities = 194/280 (69%), Positives = 229/280 (81%), Gaps = 1/280 (0%)
Frame = +1
Query: 478 DFIITSTYQEIAGSKDRPGQYESHAAFTLPGLCRVVSGINVFDPKFNIAAPGADQSIYFP 537
DFIITST+QEIAGSKD GQYESH AFTLPGL RVV GI+ FDPKFNI +PGAD SIYFP
Sbjct: 1 DFIITSTFQEIAGSKDTVGQYESHTAFTLPGLYRVVHGIDPFDPKFNIVSPGADMSIYFP 180
Query: 538 YTEKDQRHSQFHPAIEDLLFNKVDNNEHIGYLADKRKPIIFSMARLDVVKNLSGLVEWYG 597
YTE ++R ++FHP IE+LL++ V+N EHI L D+ KPIIF+MARLD VKN++GLVEWYG
Sbjct: 181 YTETERRLTEFHPDIEELLYSSVENEEHICVLKDRNKPIIFTMARLDRVKNITGLVEWYG 360
Query: 598 KNKRLRNLVNLVIVGGFFDPSK-SKDREEMAEIKKMHDLIEKYQLKGQFRWIAAQTDRYR 656
KN RLR LVNLV+V G D K SKD EE AE+KKM+ LIE Y+L GQFRWI++Q +R R
Sbjct: 361 KNARLRELVNLVVVAG--DRRKESKDLEEKAEMKKMYGLIETYKLNGQFRWISSQMNRVR 534
Query: 657 NGELYRCIADTKGAFVQPALYEAFGLTVIEAMNCGLPTFATNQGGPAEIIVDGVSGFHID 716
NGELYR I DT+GAFVQPA+YEAFGLTV+EAM CGLPTFAT GGPAEIIV G SG+HID
Sbjct: 535 NGELYRVICDTRGAFVQPAVYEAFGLTVVEAMTCGLPTFATCNGGPAEIIVHGKSGYHID 714
Query: 717 PLNGDESSNKISDFFEKCKVDPSYWNVISMAGLQRINEWY 756
P +GD ++ + +FFEK K DPS+W+ IS GL+RI+E Y
Sbjct: 715 PYHGDHAAEILVEFFEKSKADPSHWDKISQGGLKRIHEKY 834
>TC214901 homologue to UP|SUSY_SOYBN (P13708) Sucrose synthase (Sucrose-UDP
glucosyltransferase) (Nodulin-100) , partial (30%)
Length = 1013
Score = 258 bits (660), Expect = 5e-69
Identities = 127/187 (67%), Positives = 152/187 (80%), Gaps = 1/187 (0%)
Frame = +2
Query: 571 DKRKPIIFSMARLDVVKNLSGLVEWYGKNKRLRNLVNLVIVGGFFDPSK-SKDREEMAEI 629
D+ KPI+F+MAR+D V N++ LV+WYGKN +LR LVNLV+V G D K SKD EE AE+
Sbjct: 5 DRSKPIMFTMARVDRVNNITRLVDWYGKNAKLRELVNLVVVAG--DRRKESKDLEEKAEM 178
Query: 630 KKMHDLIEKYQLKGQFRWIAAQTDRYRNGELYRCIADTKGAFVQPALYEAFGLTVIEAMN 689
KKM+ LIE Y+L GQFRWI++Q +R RNGELYR I DTKGAFVQPA+YEAFGLTV+EAM
Sbjct: 179 KKMYGLIETYKLNGQFRWISSQMNRVRNGELYRVICDTKGAFVQPAIYEAFGLTVVEAMT 358
Query: 690 CGLPTFATNQGGPAEIIVDGVSGFHIDPLNGDESSNKISDFFEKCKVDPSYWNVISMAGL 749
CGLPTFAT GGPAEIIV G SGFHIDP +GD +++ + DFFEKCK+DP++W IS AGL
Sbjct: 359 CGLPTFATCNGGPAEIIVHGKSGFHIDPYHGDRAADLLVDFFEKCKLDPTHWETISKAGL 538
Query: 750 QRINEWY 756
QRI E Y
Sbjct: 539 QRIEEKY 559
>BE020760
Length = 411
Score = 226 bits (575), Expect = 4e-59
Identities = 106/136 (77%), Positives = 121/136 (88%)
Frame = +3
Query: 494 RPGQYESHAAFTLPGLCRVVSGINVFDPKFNIAAPGADQSIYFPYTEKDQRHSQFHPAIE 553
+PGQYE+H AFT+PGLCR VSGINVFDPKFNIAAPGADQS+YFP T K+QR + FHPAIE
Sbjct: 3 KPGQYETHTAFTMPGLCRAVSGINVFDPKFNIAAPGADQSVYFPSTAKEQRLTSFHPAIE 182
Query: 554 DLLFNKVDNNEHIGYLADKRKPIIFSMARLDVVKNLSGLVEWYGKNKRLRNLVNLVIVGG 613
+LL++K DN EHIG L D +KPIIFSMARLD VKNLSGLVEWY +NKRLR+LVNLV+VGG
Sbjct: 183 ELLYSKDDNEEHIGLLEDMKKPIIFSMARLDKVKNLSGLVEWYARNKRLRSLVNLVVVGG 362
Query: 614 FFDPSKSKDREEMAEI 629
FF+P+KSKDREE EI
Sbjct: 363 FFNPAKSKDREETEEI 410
>AW756812
Length = 367
Score = 161 bits (408), Expect = 9e-40
Identities = 76/121 (62%), Positives = 95/121 (77%)
Frame = +2
Query: 634 DLIEKYQLKGQFRWIAAQTDRYRNGELYRCIADTKGAFVQPALYEAFGLTVIEAMNCGLP 693
+L++KY L G FRWIAAQT+ R+GELYR +ADT+ A+VQPA YEAFGLTV+EAMNCGLP
Sbjct: 2 ELMKKYNLDGDFRWIAAQTNMARHGELYRYMADTQRAYVQPAFYEAFGLTVVEAMNCGLP 181
Query: 694 TFATNQGGPAEIIVDGVSGFHIDPLNGDESSNKISDFFEKCKVDPSYWNVISMAGLQRIN 753
TFAT GGPA+II G+SGFH+DP D++S + +F +K K DPS+W IS GLQR+
Sbjct: 182 TFATCHGGPADIIEHGMSGFHMDPYQPDQASQLLIEFIQKIKEDPSHWKKISDGGLQRMY 361
Query: 754 E 754
E
Sbjct: 362 E 364
>AI960742 homologue to GP|4098128|gb| sucrose synthase {Gossypium hirsutum},
partial (31%)
Length = 433
Score = 137 bits (346), Expect(2) = 2e-38
Identities = 65/111 (58%), Positives = 84/111 (75%)
Frame = +3
Query: 228 YQKFELRLKEWGFEKGWGDNAGRVKETMRTLSEVLQAPDPVNLEIFFSRIPTIFKVVIFS 287
Y +FE + +E G E+GWGD A RV E ++ L ++L+APDP LE F R+P +F VVI S
Sbjct: 3 YSEFENKFREIGLERGWGDIAERVLEMIQLLLDLLEAPDPCTLETFLGRVPMVFNVVILS 182
Query: 288 VHGYFGQADVLGLPDTGGQVVYILDQVKALEEELILRIKQQGLNYKPQILV 338
HGYF Q +VLG PDTGGQVVYILDQV+ALE E++ RIK+QGL+ P+IL+
Sbjct: 183 PHGYFAQDNVLGYPDTGGQVVYILDQVRALENEMLNRIKKQGLDITPRILI 335
Score = 40.8 bits (94), Expect(2) = 2e-38
Identities = 18/31 (58%), Positives = 22/31 (70%)
Frame = +2
Query: 348 GTKCHQEFEPINDTKHSHILRVPFHTEKGIL 378
GT C Q E + DT++ ILRVPF TEKGI+
Sbjct: 338 GTTCGQRLERVYDTEYCDILRVPFRTEKGIV 430
>BI971794 homologue to SP|P13708|SUSY Sucrose synthase (EC 2.4.1.13)
(Sucrose-UDP glucosyltransferase) (Nodulin-100).
[Soybean], partial (13%)
Length = 345
Score = 85.5 bits (210), Expect(2) = 4e-35
Identities = 36/62 (58%), Positives = 48/62 (77%)
Frame = +2
Query: 341 RLIPDARGTKCHQEFEPINDTKHSHILRVPFHTEKGILPQWVSRFDIYPYLERFTQDATT 400
RL+PDA GT C Q E + T+HSHILRVPF TEKGI+ QW+SRF+++PYLE +T+D
Sbjct: 2 RLLPDAIGTTCGQRLEKVFGTEHSHILRVPFRTEKGIVRQWISRFEVWPYLETYTEDVAH 181
Query: 401 KI 402
++
Sbjct: 182 EL 187
Score = 82.0 bits (201), Expect(2) = 4e-35
Identities = 36/48 (75%), Positives = 43/48 (89%)
Frame = +1
Query: 408 GKPDLVIGNYTDGNLVASLMARKLGITQATIAHALEKTKYEDSDVKWK 455
GKPDL+ GNY+DGN+VASL+A KLG+TQ TIAHALEKTKY +SD+ WK
Sbjct: 202 GKPDLIGGNYSDGNIVASLLAHKLGVTQCTIAHALEKTKYPESDIYWK 345
>TC225395 homologue to UP|SUSY_SOYBN (P13708) Sucrose synthase (Sucrose-UDP
glucosyltransferase) (Nodulin-100) , partial (18%)
Length = 578
Score = 141 bits (355), Expect = 1e-33
Identities = 65/89 (73%), Positives = 75/89 (84%)
Frame = +3
Query: 668 KGAFVQPALYEAFGLTVIEAMNCGLPTFATNQGGPAEIIVDGVSGFHIDPLNGDESSNKI 727
KGAFVQPA+YEAFGLTV+EAM CGLPTFAT GGPAEIIV G SGFHIDP +GD +++ +
Sbjct: 3 KGAFVQPAIYEAFGLTVVEAMTCGLPTFATCNGGPAEIIVHGKSGFHIDPYHGDRAADLL 182
Query: 728 SDFFEKCKVDPSYWNVISMAGLQRINEWY 756
DFFEKCK+DP++W IS AGLQRI E Y
Sbjct: 183 VDFFEKCKLDPTHWETISKAGLQRIEEKY 269
>TC225851 similar to UP|SUS2_PEA (O24301) Sucrose synthase 2 (Sucrose-UDP
glucosyltransferase 2) , partial (19%)
Length = 641
Score = 134 bits (336), Expect = 2e-31
Identities = 68/106 (64%), Positives = 78/106 (73%)
Frame = +2
Query: 660 LYRCIADTKGAFVQPALYEAFGLTVIEAMNCGLPTFATNQGGPAEIIVDGVSGFHIDPLN 719
LYR IADT AFVQ A YEAFGLTV+EAMNCGLPTFAT GGPAEII G+SGFHIDP +
Sbjct: 2 LYRYIADTXXAFVQXAXYEAFGLTVVEAMNCGLPTFATCHGGPAEIIEHGISGFHIDPYH 181
Query: 720 GDESSNKISDFFEKCKVDPSYWNVISMAGLQRINEWYYLRPLLRCF 765
D++S + +FF+K K DPS+W IS GLQRI E + L CF
Sbjct: 182 PDQASQLLVEFFQKSKEDPSHWKKISDGGLQRIYER*VILLKLCCF 319
>TC225852 similar to UP|Q9SLY2 (Q9SLY2) Sucrose synthase, partial (12%)
Length = 740
Score = 133 bits (335), Expect = 3e-31
Identities = 66/99 (66%), Positives = 76/99 (76%)
Frame = +1
Query: 492 KDRPGQYESHAAFTLPGLCRVVSGINVFDPKFNIAAPGADQSIYFPYTEKDQRHSQFHPA 551
K+ GQYESHA FTLPGL RVV GI+VFDPKFNI +PGAD SIYFPY+EK R + H +
Sbjct: 442 KNTVGQYESHAGFTLPGLYRVVHGIDVFDPKFNIVSPGADMSIYFPYSEKQNRLTALHGS 621
Query: 552 IEDLLFNKVDNNEHIGYLADKRKPIIFSMARLDVVKNLS 590
IE LLF +E+IG L DK KPIIFSMARLD VKN++
Sbjct: 622 IEQLLFAPEQTDEYIGLLKDKSKPIIFSMARLDRVKNIT 738
>BM108319 homologue to SP|P13708|SUSY_ Sucrose synthase (EC 2.4.1.13)
(Sucrose-UDP glucosyltransferase) (Nodulin-100).
[Soybean], partial (10%)
Length = 440
Score = 113 bits (283), Expect = 3e-25
Identities = 58/119 (48%), Positives = 74/119 (61%)
Frame = +1
Query: 243 GWGDNAGRVKETMRTLSEVLQAPDPVNLEIFFSRIPTIFKVVIFSVHGYFGQADVLGLPD 302
GWGDNA RV E+++ L ++L+APDP LE F RIP +F VVI S HGYF Q +VLG PD
Sbjct: 1 GWGDNAXRVLESIQLLLDLLEAPDPCTLETFLGRIPMVFNVVILSPHGYFAQDNVLGYPD 180
Query: 303 TGGQVVYILDQVKALEEELILRIKQQGLNYKPQILVVTRLIPDARGTKCHQEFEPINDT 361
TGGQ VYILDQ +AL ++ I + + L++T P GT C Q E + T
Sbjct: 181 TGGQFVYILDQFRALXNXMLHPINNK-IGXGTXFLIITXSPPXCNGTTCGQFLEXVFGT 354
>TC210502 homologue to UP|Q9AXK3 (Q9AXK3) Sucrose-phosphate synthase ,
partial (30%)
Length = 966
Score = 112 bits (280), Expect = 6e-25
Identities = 93/326 (28%), Positives = 149/326 (45%), Gaps = 8/326 (2%)
Frame = +3
Query: 424 ASLMARKLGITQATIAHALEKTKYED----SDVKWKELDPKYHFSCQFMADTVAMNSSDF 479
A+L++ L + H+L + K E + E++ Y + A+ +A++ S+
Sbjct: 12 AALLSGALNVPMLFTGHSLGRDKLEQLLKQGRLSKDEINTTYKIMRRIEAEELALDGSEI 191
Query: 480 IITSTYQEIAGSKDRPGQYESHAAFTLPG-LCRVVSGINVFDPKFNIAAPGADQSIYFPY 538
+ITST QEI ++ L + R VS F P+ PG + P+
Sbjct: 192 VITSTRQEIEEQWRLYDGFDPVLERKLRARIRRNVSCYGRFMPRMATIPPGMEFHHIVPH 371
Query: 539 T---EKDQRHSQFHPAIEDLLFNKVDNNEHIGYLADKRKPIIFSMARLDVVKNLSGLVEW 595
E + + HPA +D +E + + + RKP+I ++AR D KN++ LV+
Sbjct: 372 DGDIEGEPEGNLDHPAPQDPPIW----SEIMRFFTNPRKPMILALARPDPKKNITTLVKA 539
Query: 596 YGKNKRLRNLVNLVIVGGFFDPSKSKDREEMAEIKKMHDLIEKYQLKGQFRWIAAQTDRY 655
+G+ + L+ L NL ++ G D + + + LI+KY L GQ + +Y
Sbjct: 540 FGECRPLQELANLTLIMGNRDGIDEMSSTNASVLLSVLKLIDKYDLYGQVAY-PKHHKQY 716
Query: 656 RNGELYRCIADTKGAFVQPALYEAFGLTVIEAMNCGLPTFATNQGGPAEIIVDGVSGFHI 715
++YR A TKG F+ PA E FGLT+IEA GLP AT GGP +I +G +
Sbjct: 717 DVPDIYRLAAKTKGVFINPAFIEPFGLTLIEAAAHGLPIVATKNGGPVDIHRVLDNGLLV 896
Query: 716 DPLNGDESSNKISDFFEKCKVDPSYW 741
DP I+D K + W
Sbjct: 897 DP----HDQQSIADALLKLVSNKQLW 962
>BE556484 similar to SP|P13708|SUSY_ Sucrose synthase (EC 2.4.1.13)
(Sucrose-UDP glucosyltransferase) (Nodulin-100).
[Soybean], partial (9%)
Length = 238
Score = 101 bits (251), Expect = 1e-21
Identities = 48/79 (60%), Positives = 58/79 (72%)
Frame = +2
Query: 458 DPKYHFSCQFMADTVAMNSSDFIITSTYQEIAGSKDRPGQYESHAAFTLPGLCRVVSGIN 517
+ +YHF CQF AD AMN +D II+ST+QEIAGSKD GQ E AF L GL RV GI+
Sbjct: 2 EERYHFFCQFTADLFAMNHTDLIISSTFQEIAGSKDTVGQCEFDTAFALTGLYRVAHGID 181
Query: 518 VFDPKFNIAAPGADQSIYF 536
VFDPK N+ +PGA+ +IYF
Sbjct: 182 VFDPKLNMVSPGAESTIYF 238
>TC232827 similar to UP|Q94G60 (Q94G60) Sucrose synthase , partial (8%)
Length = 899
Score = 95.1 bits (235), Expect = 1e-19
Identities = 45/64 (70%), Positives = 55/64 (85%)
Frame = +1
Query: 275 SRIPTIFKVVIFSVHGYFGQADVLGLPDTGGQVVYILDQVKALEEELILRIKQQGLNYKP 334
+R+P +F +VI + GYFGQA VLGLPDTGGQVVYILDQV+ALEEEL+ +I+ QGL+ KP
Sbjct: 22 TRLPNMFNIVILCIRGYFGQAAVLGLPDTGGQVVYILDQVRALEEELLHKIELQGLDVKP 201
Query: 335 QILV 338
QILV
Sbjct: 202 QILV 213
>BG507656 similar to SP|P13708|SUSY_ Sucrose synthase (EC 2.4.1.13)
(Sucrose-UDP glucosyltransferase) (Nodulin-100).
[Soybean], partial (9%)
Length = 280
Score = 90.1 bits (222), Expect = 3e-18
Identities = 50/82 (60%), Positives = 60/82 (72%), Gaps = 1/82 (1%)
Frame = +2
Query: 560 VDNNEHIGYLADKRKPIIFSMARLDVVKNLSGLVEWYGKNKRLRNLVNLVIVGGFFDPSK 619
++N EHI L D+ KPIIF+MARLD VKN++GLVEW+ KN RLR LVNLV+V G D K
Sbjct: 5 LENEEHICVLKDRIKPIIFTMARLDRVKNITGLVEWFFKNARLRKLVNLVVVAG--DRXK 178
Query: 620 S-KDREEMAEIKKMHDLIEKYQ 640
KD EE A +KKM+ LIE Q
Sbjct: 179 DPKDLEEXAAMKKMYGLIEYLQ 244
>AI965972 similar to SP|P13708|SUSY_ Sucrose synthase (EC 2.4.1.13)
(Sucrose-UDP glucosyltransferase) (Nodulin-100).
[Soybean], partial (10%)
Length = 250
Score = 89.4 bits (220), Expect = 6e-18
Identities = 42/70 (60%), Positives = 55/70 (78%)
Frame = +2
Query: 619 KSKDREEMAEIKKMHDLIEKYQLKGQFRWIAAQTDRYRNGELYRCIADTKGAFVQPALYE 678
+SKD E+ +KKM+ LIE Y+L GQ+RWI++Q +R RNG+LYR I DTKGAFVQ A+Y+
Sbjct: 41 ESKDLEDKDVMKKMYGLIETYKLNGQYRWISSQMNRARNGKLYRVICDTKGAFVQTAIYK 220
Query: 679 AFGLTVIEAM 688
A GLT EA+
Sbjct: 221 AIGLTAAEAI 250
>TC225853 homologue to UP|Q9SLY2 (Q9SLY2) Sucrose synthase, partial (8%)
Length = 419
Score = 88.2 bits (217), Expect = 1e-17
Identities = 41/60 (68%), Positives = 47/60 (78%)
Frame = -2
Query: 490 GSKDRPGQYESHAAFTLPGLCRVVSGINVFDPKFNIAAPGADQSIYFPYTEKDQRHSQFH 549
G+K+ QYESHA FTLPGL RVV GI+VFDPKFNI +PGAD SIYFPY+EK R + H
Sbjct: 187 GTKNTVVQYESHAGFTLPGLYRVVHGIDVFDPKFNIVSPGADMSIYFPYSEKQNRLTALH 8
>TC222890 similar to UP|SUS2_PEA (O24301) Sucrose synthase 2 (Sucrose-UDP
glucosyltransferase 2) , partial (14%)
Length = 425
Score = 87.8 bits (216), Expect = 2e-17
Identities = 46/114 (40%), Positives = 67/114 (58%)
Frame = +2
Query: 1 MAPTHALKRTNSIADNMPDALRKSRYHMKKCFAKYLEKGRRIMKLHELMEEVERTIDDIN 60
M+ L R SI D + D L R + ++Y+ +GR I++ H LM+E++ D
Sbjct: 80 MSTQPKLGRIPSIRDRVEDTLSAHRNELISLLSRYVAQGRGILQPHNLMDELDNIPGDDQ 259
Query: 61 ERNYILEGNLGFILSSTQEAVVDPPYVAFAIRPNPGVWEYVRVNSEDLSVEPIT 114
+ G G I+ S +EA+V PP+VA A+RP PGVWEYVRVN +LSVE ++
Sbjct: 260 AIVDLKNGPFGEIVKSAKEAIVLPPFVAIAVRPRPGVWEYVRVNVSELSVEQLS 421
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.138 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 31,733,929
Number of Sequences: 63676
Number of extensions: 408390
Number of successful extensions: 1732
Number of sequences better than 10.0: 56
Number of HSP's better than 10.0 without gapping: 1714
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1723
length of query: 780
length of database: 12,639,632
effective HSP length: 105
effective length of query: 675
effective length of database: 5,953,652
effective search space: 4018715100
effective search space used: 4018715100
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC148342.7


