

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148291.4 + phase: 0 /pseudo
(659 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
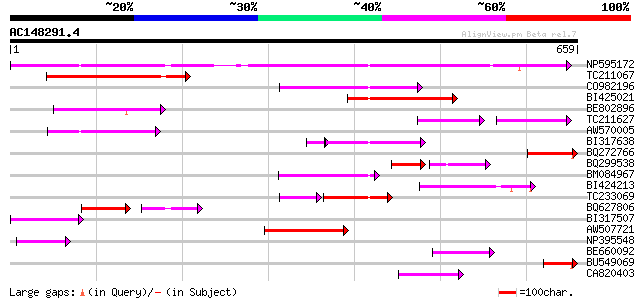
Score E
Sequences producing significant alignments: (bits) Value
NP595172 polyprotein [Glycine max] 424 e-119
TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%) 139 3e-33
CO982196 123 3e-28
BI425021 118 7e-27
BE802896 99 6e-21
TC211627 71 3e-20
AW570005 93 4e-19
BI317638 weakly similar to GP|9294238|dbj| contains similarity t... 86 4e-18
BQ272766 weakly similar to GP|28558781|gb| pol protein {Cucumis ... 87 2e-17
BQ299538 54 7e-17
BM084967 85 1e-16
BI424213 82 8e-16
TC233069 69 5e-14
BQ627806 48 2e-13
BI317507 70 2e-12
AW507721 weakly similar to GP|27764548|gb polyprotein {Glycine m... 70 2e-12
NP395548 reverse transcriptase [Glycine max] 54 2e-07
BE660092 weakly similar to GP|9884624|dbj retroelement pol polyp... 53 5e-07
BU549069 52 1e-06
CA820403 weakly similar to GP|13273463|gb| pol protein integrase... 51 2e-06
>NP595172 polyprotein [Glycine max]
Length = 4659
Score = 424 bits (1089), Expect = e-119
Identities = 248/657 (37%), Positives = 364/657 (54%), Gaps = 4/657 (0%)
Frame = +1
Query: 1 RLYAKLSKCEFWLSEVSFLGHIIFGSDIAVDPSKVDAVSQWETLKSVTEIRSFLGLAGYY 60
+L+A+LSKC F +EV +LGH + G ++++ +KV AV W T +V ++R FLGL GYY
Sbjct: 2332 QLFARLSKCSFGDTEVDYLGHKVSGLGVSMENTKVQAVLDWPTPNNVKQLRGFLGLTGYY 2511
Query: 61 RRFIEGFSKLALPSTQLTCKGKSFVWDAQCERSFNELKRRLTIAPVLILPKPEEPFVVYC 120
RRFI+ ++ +A P T L K SF+W+ + E +F +LK+ +T APVL LP +PF++
Sbjct: 2512 RRFIKSYANIAGPLTDLLQKD-SFLWNNEAEAAFVKLKKAMTEAPVLSLPDFSQPFILET 2688
Query: 121 DASKLGLGGVLMQDGKVVAYASRQLRVHEKNYPTHDLELAAVVFVLKIWRHYLYGSRFEV 180
DAS +G+G VL Q+G +AY S++L + + EL A+ L +RHYL G++F +
Sbjct: 2689 DASGIGVGAVLGQNGHPIAYFSKKLAPRMQKQSAYTRELLAITEALSKFRHYLLGNKFII 2868
Query: 181 FRFEVFSDHKSLKYLFDQKELNMRQRRWLELLKDYDFGLNYHPG*ANVVADALSRKTLHM 240
+D +SLK L DQ Q+ WL YDF + Y PG N ADALSR +
Sbjct: 2869 R-----TDQRSLKSLMDQSLQTPEQQAWLHKFLGYDFKIEYKPGKDNQAADALSRMFM-- 3027
Query: 241 SALMVKEFELLEHFRDMSLVCELSFQSIHLGMLKIDSDFLNSIREAQKVDLKFVDLMTSG 300
L++ H S FL +R D LM +
Sbjct: 3028 ----------------------LAWSEPH-------SIFLEELRARLISDPHLKQLMETY 3120
Query: 301 NDTEDSDFKVDDQGVLRFRGKVCIPDNEELKKLILEESHKSRLSIHPGATKMYHDLKKLF 360
D+ +G+L ++ +V IP EE+ IL+E H S + H G T+ LK F
Sbjct: 3121 KQGADASHYTVREGLLYWKDRVVIPAEEEIVNKILQEYHSSPIGGHAGITRTLARLKAQF 3300
Query: 361 WWSGLKRDVAQFVYACLICQKSKVEHQKPAGLLTPLDVPEWKWDSISMDFVTSLPNTPRG 420
+W ++ DV ++ CLICQ++K + PAGLL PL +P+ W+ ++MDF+T LPN+ G
Sbjct: 3301 YWPKMQEDVKAYIQKCLICQQAKSNNTLPAGLLQPLPIPQQVWEDVAMDFITGLPNS-FG 3477
Query: 421 HDAIWVVVDRLTKSAHFIPININYPVAQLAEIYIHSVVKLHGVPASIVSDRDPRFTSRFW 480
I VV+DRLTK AHFIP+ +Y +AE ++ +VKLHG+P SIVSDRD FTS FW
Sbjct: 3478 LSVIMVVIDRLTKYAHFIPLKADYNSKVVAEAFMSHIVKLHGIPRSIVSDRDRVFTSTFW 3657
Query: 481 KRLQDALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGGAWDSHLPLIEFTYNN 540
+ L G+ L +SSAYHPQ+DGQSE + LE LR E W LP EF YN
Sbjct: 3658 QHLFKLQGTTLAMSSAYHPQSDGQSEVLNKCLEMYLRCFTYEHPKGWVKALPWAEFWYNT 3837
Query: 541 SYHSSIGMAPFEALYGRRCRTPLCWFESGESVVLGPDLVHETTEKVRLIRE---RMKASQ 597
+YH S+GM PF ALYGR P S+ ++ + T++ L+ + + +Q
Sbjct: 3838 AYHMSLGMTPFRALYGRE---PPTLTRQACSIDDPAEVREQLTDRDALLAKLKINLTRAQ 4008
Query: 598 SRQKSYHDKRRKELEFQEGDHVFLRVTPLTGVGRAL-KSRKLTPKFIGPYQISDRVG 653
K DK+R ++ FQ GD V +++ P L K++KL+ ++ GP+++ ++G
Sbjct: 4009 QVMKRQADKKRLDVSFQIGDEVLVKLQPYRQHSAVLRKNQKLSMRYFGPFKVLAKIG 4179
>TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%)
Length = 589
Score = 139 bits (351), Expect = 3e-33
Identities = 74/168 (44%), Positives = 103/168 (61%)
Frame = +1
Query: 43 TLKSVTEIRSFLGLAGYYRRFIEGFSKLALPSTQLTCKGKSFVWDAQCERSFNELKRRLT 102
TLKSV +IRSF GLA +YRRF+ FS +A P +L K +F W + E++F LK +LT
Sbjct: 100 TLKSVGDIRSFHGLASFYRRFVPNFSTVASPLNELVKKNMAFTWGEKQEQAFALLKEKLT 279
Query: 103 IAPVLILPKPEEPFVVYCDASKLGLGGVLMQDGKVVAYASRQLRVHEKNYPTHDLELAAV 162
APVL LP + F + CDAS +G+ VL+Q G +AY S +L NYPT+D EL A+
Sbjct: 280 KAPVLALPDFSKTFELECDASGVGVRAVLLQGGHPIAYFSEKLHSATLNYPTYDKELYAL 459
Query: 163 VFVLKIWRHYLYGSRFEVFRFEVFSDHKSLKYLFDQKELNMRQRRWLE 210
+ + W H+L F + SDH+SLKY+ + +LN R +W+E
Sbjct: 460 IRAPQTWEHFLVCK-----EFVIHSDHQSLKYIRGKSKLNKRHAKWVE 588
Score = 29.3 bits (64), Expect = 5.8
Identities = 19/76 (25%), Positives = 35/76 (46%)
Frame = +2
Query: 11 FWLSEVSFLGHIIFGSDIAVDPSKVDAVSQWETLKSVTEIRSFLGLAGYYRRFIEGFSKL 70
F + + F G ++ + + +DP K+ A+ +W V EI LG + + IEG +
Sbjct: 2 FCVDNIFFSGFVVGRNGVQMDPEKIKAIQEWPPP*KVWEI---LGASMG*QASIEGSFLI 172
Query: 71 ALPSTQLTCKGKSFVW 86
+L L+ +W
Sbjct: 173SLQLHHLSMSW*RRIW 220
>CO982196
Length = 812
Score = 123 bits (308), Expect = 3e-28
Identities = 67/166 (40%), Positives = 95/166 (56%)
Frame = +1
Query: 314 GVLRFRGKVCIPDNEELKKLILEESHKSRLSIHPGATKMYHDLKKLFWWSGLKRDVAQFV 373
G L F+ ++ + N L+L+E S L H G + + + + +W G+K+ +V
Sbjct: 316 GKLYFKDRLVLSKNSTKIPLLLKELQDSPLGGHSGFFRTFKRVANVVFWQGMKKTTRDYV 495
Query: 374 YACLICQKSKVEHQKPAGLLTPLDVPEWKWDSISMDFVTSLPNTPRGHDAIWVVVDRLTK 433
AC IC+++K PAGLL L +P W ISMDF+ LP +G D I VVVDRLTK
Sbjct: 496 AACEICRRNKTSTLSPAGLL*LLPIPTKVWTDISMDFIGGLPKA-QGKDNILVVVDRLTK 672
Query: 434 SAHFIPININYPVAQLAEIYIHSVVKLHGVPASIVSDRDPRFTSRF 479
AHF ++ Y ++AE++I +V+LHG PASIVSD F S F
Sbjct: 673 YAHFFALSHPYTAKEVAELFIKELVRLHGFPASIVSDXXRLFMSLF 810
>BI425021
Length = 426
Score = 118 bits (296), Expect = 7e-27
Identities = 60/128 (46%), Positives = 82/128 (63%)
Frame = -1
Query: 393 LTPLDVPEWKWDSISMDFVTSLPNTPRGHDAIWVVVDRLTKSAHFIPININYPVAQLAEI 452
L PL VP+ W+ +SMDF+ LP GH I+VVV+R +K H + ++ +A +
Sbjct: 426 LCPLPVPQRPWEDLSMDFIVGLPPY-HGHTTIFVVVNRFSKGIHLGTLPTSHTAHMVASL 250
Query: 453 YIHSVVKLHGVPASIVSDRDPRFTSRFWKRLQDALGSKLRLSSAYHPQTDGQSERTIQSL 512
+++ V+KLHG P SIVSDRDP F S FW+ L G+ LR+SSAYHPQTDGQ+E + +
Sbjct: 249 FLNIVIKLHGFPRSIVSDRDPLFISHFWQDLFRLSGTVLRMSSAYHPQTDGQTEVLNRVI 70
Query: 513 EDLLRVCV 520
E LR V
Sbjct: 69 EQYLRAFV 46
>BE802896
Length = 416
Score = 99.0 bits (245), Expect = 6e-21
Identities = 57/134 (42%), Positives = 79/134 (58%), Gaps = 4/134 (2%)
Frame = -2
Query: 52 SFLGLAGYYRRFIEGFSKLALPSTQLTCKGKSFVWDAQCERSFNELKRRLTIAPVLILPK 111
SFLG AG+YRRFI F K+ALP + L K F ++ +C+ +F+ LKR L P++ P
Sbjct: 415 SFLGHAGFYRRFIRDFRKVALPLSNLLQKEVEFDFNDKCK*AFDCLKRALITTPIIQAPD 236
Query: 112 PEEPFVVYCDASKLGLGGVLMQD----GKVVAYASRQLRVHEKNYPTHDLELAAVVFVLK 167
PF + CDAS LG VL Q +V+ Y+SR L + NY T + EL A+VF L+
Sbjct: 235 WTAPFELMCDASNYALGVVLAQKIDKLPRVIYYSSRTLDAAQANYTTTEKELLAIVFALE 56
Query: 168 IWRHYLYGSRFEVF 181
+ YL G+R V+
Sbjct: 55 KFHSYLLGTRIIVY 14
>TC211627
Length = 1034
Score = 71.2 bits (173), Expect(2) = 3e-20
Identities = 38/78 (48%), Positives = 44/78 (55%)
Frame = +2
Query: 475 FTSRFWKRLQDALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGGAWDSHLPLI 534
F S W L G+KLR S+AYHPQTDGQ+E + LE LR V + W L L
Sbjct: 5 FISGLWHELFHISGTKLRFSTAYHPQTDGQTEVINRILEQYLRAFVHDHPQHWFKFLSLA 184
Query: 535 EFTYNNSYHSSIGMAPFE 552
E YN S HS IG +PFE
Sbjct: 185 E*CYNTSVHSGIGFSPFE 238
Score = 45.8 bits (107), Expect(2) = 3e-20
Identities = 28/89 (31%), Positives = 42/89 (46%), Gaps = 1/89 (1%)
Frame = +3
Query: 566 FESGESVVLGPD-LVHETTEKVRLIRERMKASQSRQKSYHDKRRKELEFQEGDHVFLRVT 624
+ +G S V D ++H + R++ Q K D R++L F GD V++R+
Sbjct: 276 YSAGTSTVEAVDAILHSLATIHHTLTCRLQKYQDSMKRIADSHRRDLTFNIGDWVYVRL* 455
Query: 625 PLTGVGRALKSRKLTPKFIGPYQISDRVG 653
P KL+ +F GPYQI RVG
Sbjct: 456 PYRQTSIQSTYTKLSKRFYGPYQIQARVG 542
>AW570005
Length = 413
Score = 92.8 bits (229), Expect = 4e-19
Identities = 52/131 (39%), Positives = 75/131 (56%)
Frame = -2
Query: 45 KSVTEIRSFLGLAGYYRRFIEGFSKLALPSTQLTCKGKSFVWDAQCERSFNELKRRLTIA 104
++ +R FL L G+YRRFI+G++ +A P + L K SFVW + + +F LK +T
Sbjct: 403 RTARSLRGFLRLTGFYRRFIKGYAAMAAPLSHLLTKD-SFVWSPEADVAFQALKNVVTNT 227
Query: 105 PVLILPKPEEPFVVYCDASKLGLGGVLMQDGKVVAYASRQLRVHEKNYPTHDLELAAVVF 164
VL LP +PF V DAS +G VL Q+G +A+ S++ T+ ELAA+
Sbjct: 226 LVLALPDFTKPFTVETDASGSDMGAVLSQEGHPIAFFSKEFCPKLVRSSTYVHELAAITN 47
Query: 165 VLKIWRHYLYG 175
V+K WR YL G
Sbjct: 46 VVKKWRQYLLG 14
>BI317638 weakly similar to GP|9294238|dbj| contains similarity to reverse
transcriptase~gene_id:K11J14.5 {Arabidopsis thaliana},
partial (5%)
Length = 420
Score = 86.3 bits (212), Expect(2) = 4e-18
Identities = 48/116 (41%), Positives = 67/116 (57%)
Frame = -2
Query: 368 DVAQFVYACLICQKSKVEHQKPAGLLTPLDVPEWKWDSISMDFVTSLPNTPRGHDAIWVV 427
+ AQ + CL CQ +K E ++ LL PL VP W+ +S+DF+T L H AI VV
Sbjct: 350 ECAQMLPNCLDCQHTKYETKRIVDLLCPLLVPHRPWEDLSLDFITGLLPY-HVHTAILVV 174
Query: 428 VDRLTKSAHFIPININYPVAQLAEIYIHSVVKLHGVPASIVSDRDPRFTSRFWKRL 483
VD +K H + ++ +A ++I SV KLHG+P S+VSD D F S FW+ L
Sbjct: 173 VDHFSKGIHLGMLPSSHTAHTVACLFIDSVAKLHGLPRSLVSDCDLLFVSHFWQEL 6
Score = 23.9 bits (50), Expect(2) = 4e-18
Identities = 10/26 (38%), Positives = 13/26 (49%)
Frame = -1
Query: 346 HPGATKMYHDLKKLFWWSGLKRDVAQ 371
H G K L K +W G++ DV Q
Sbjct: 405 HTGIAKTLA*LSKNIYWFGMRTDVTQ 328
>BQ272766 weakly similar to GP|28558781|gb| pol protein {Cucumis melo},
partial (9%)
Length = 410
Score = 87.0 bits (214), Expect = 2e-17
Identities = 45/65 (69%), Positives = 50/65 (76%), Gaps = 7/65 (10%)
Frame = -3
Query: 602 SYHDKRRKELEFQEGDHVFLRVTPLTGVGRALKSRKLTPKFIGPYQISDRV-------GL 654
SYHDKRRK+LEF+ GDHVFLRVTP TGVGRALKS KLTP FIGP+QI +V L
Sbjct: 408 SYHDKRRKDLEFEVGDHVFLRVTP*TGVGRALKS*KLTPHFIGPFQILKKVDFVAYQIAL 229
Query: 655 PPHLS 659
PP L+
Sbjct: 228 PPSLT 214
>BQ299538
Length = 426
Score = 53.5 bits (127), Expect(2) = 7e-17
Identities = 28/71 (39%), Positives = 41/71 (57%)
Frame = +3
Query: 488 GSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGGAWDSHLPLIEFTYNNSYHSSIG 547
G+ L++S++YHP DGQ+ LE LR V +Q L E+ YN ++H+S G
Sbjct: 144 GTYLKMSTSYHP*IDGQTVN--HCLETFLRCFVADQPKM*VQWLSWAEYWYNTNFHASTG 317
Query: 548 MAPFEALYGRR 558
PFE +YGR+
Sbjct: 318 TTPFEVVYGRK 350
Score = 52.4 bits (124), Expect(2) = 7e-17
Identities = 24/40 (60%), Positives = 27/40 (67%)
Frame = +1
Query: 444 YPVAQLAEIYIHSVVKLHGVPASIVSDRDPRFTSRFWKRL 483
Y LAEI+ VV LHGVPAS++SD DP F S FWK L
Sbjct: 13 YSARVLAEIFTKEVVHLHGVPASVLSDEDPIFVSSFWKEL 132
>BM084967
Length = 426
Score = 84.7 bits (208), Expect = 1e-16
Identities = 49/118 (41%), Positives = 64/118 (53%)
Frame = -2
Query: 313 QGVLRFRGKVCIPDNEELKKLILEESHKSRLSIHPGATKMYHDLKKLFWWSGLKRDVAQF 372
Q ++ G + +P +L E H S H G TK L + F W G+++DV QF
Sbjct: 356 QDLILKNGCIWLPSGFSFIPTLLLEYHSSPTDAHIGVTKTMARLSENFTWIGIRKDVEQF 177
Query: 373 VYACLICQKSKVEHQKPAGLLTPLDVPEWKWDSISMDFVTSLPNTPRGHDAIWVVVDR 430
V ACL CQ +K E QK AGLL PL VP W+ +S +F+ L RG+ AI VVV R
Sbjct: 176 VAACLDCQYTKYEAQKMAGLLCPLPVPCRPWEDLSFNFIIGLSEF-RGYTAILVVVGR 6
>BI424213
Length = 426
Score = 82.0 bits (201), Expect = 8e-16
Identities = 51/143 (35%), Positives = 73/143 (50%), Gaps = 8/143 (5%)
Frame = +1
Query: 477 SRFWKRLQDALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGGAWDSHLPLIEF 536
S FWK L LG+KL S+ HPQTDGQ++ +SL LLR + +WD +LP +EF
Sbjct: 4 SHFWKTLWAKLGTKLLFSTTCHPQTDGQTKVVNRSLSTLLRALLKGNHKSWDEYLPHVEF 183
Query: 537 TYNNSYHSSIGMAPFEALYGRRCRTPLCWFESGESVVLGPDLVHE----TTEKVRLIRER 592
YN H + +PFE +YG TPL + L +H+ +E V+ + ER
Sbjct: 184 AYNRGVHRTTKQSPFEVVYGFNPLTPLDLI----PLPLDTSFIHKEGESRSEFVKKMHER 351
Query: 593 MKASQSRQKSYH----DKRRKEL 611
+K Q + ++ RKEL
Sbjct: 352 VKNQIENQTKVYSTKGNRGRKEL 420
>TC233069
Length = 881
Score = 69.3 bits (168), Expect(2) = 5e-14
Identities = 38/82 (46%), Positives = 50/82 (60%), Gaps = 2/82 (2%)
Frame = +2
Query: 365 LKRDVAQFVYACLICQKSKVEHQKPAGLLTPLDVPEWKWDSISMDFVTSLPNTPRGHDAI 424
+ RDV + AC CQ++K QK GLL PL +P W ISMDFVT LP + G I
Sbjct: 224 MARDVCDHICACTNCQQNKYSTQK*FGLLQPLPIP*QVWKDISMDFVTHLPPS-*GKKMI 400
Query: 425 WVVVDRLTKSAHF--IPININY 444
WV+VD TK +HF +P +++Y
Sbjct: 401 WVIVDCWTKYSHFLSLPAHLSY 466
Score = 26.9 bits (58), Expect(2) = 5e-14
Identities = 16/49 (32%), Positives = 23/49 (46%)
Frame = +1
Query: 314 GVLRFRGKVCIPDNEELKKLILEESHKSRLSIHPGATKMYHDLKKLFWW 362
G+L + + IP EL+ ++L E H S L H G + L F W
Sbjct: 73 GLLLYCTHLFIPLESELRAILLGECHDSLLGGHSGIKGTLNRLFVSFAW 219
>BQ627806
Length = 435
Score = 48.1 bits (113), Expect(2) = 2e-13
Identities = 24/57 (42%), Positives = 36/57 (63%)
Frame = +3
Query: 84 FVWDAQCERSFNELKRRLTIAPVLILPKPEEPFVVYCDASKLGLGGVLMQDGKVVAY 140
F W+ + +R+F++LK L APVL LP FVV DAS +G+G +L Q+ +A+
Sbjct: 21 FHWNEEADRAFSQLKLALCQAPVLGLPDFNSSFVVETDASGIGMGAILSQNHHPLAF 191
Score = 46.2 bits (108), Expect(2) = 2e-13
Identities = 25/71 (35%), Positives = 36/71 (50%)
Frame = +1
Query: 154 THDLELAAVVFVLKIWRHYLYGSRFEVFRFEVFSDHKSLKYLFDQKELNMRQRRWLELLK 213
T+ ELAA+ +K WR YL G F + +DH+SLK L Q Q+ +L L
Sbjct: 232 TYVRELAAITVAVKKWRQYLLGHHFVIL-----TDHRSLKELMSQAVQTPEQQIYLARLM 396
Query: 214 DYDFGLNYHPG 224
+D+ + Y G
Sbjct: 397 GFDYTIQYRAG 429
>BI317507
Length = 359
Score = 70.5 bits (171), Expect = 2e-12
Identities = 38/87 (43%), Positives = 52/87 (59%), Gaps = 1/87 (1%)
Frame = -1
Query: 1 RLYAKLSKCEFWLSEVSFLGHIIFGSDIAVDPSKVDAVSQWETLKSVTEIRSFLGLAGYY 60
RL A KC F + + +LGH+I +A+D +KV +V +W K+V + SFL L GYY
Sbjct: 275 RLVANRKKCYFSQTTIEYLGHVISKDCVAMDSNKVKSVIEWPVPKNVKRVCSFLRLTGYY 96
Query: 61 RRFIEGFSKLA-LPSTQLTCKGKSFVW 86
R+FI+ + KLA P T LT K F W
Sbjct: 95 RKFIKDYGKLAPRPLTDLT-KNDGFKW 18
>AW507721 weakly similar to GP|27764548|gb polyprotein {Glycine max}, partial
(3%)
Length = 464
Score = 70.5 bits (171), Expect = 2e-12
Identities = 34/98 (34%), Positives = 60/98 (60%), Gaps = 1/98 (1%)
Frame = +1
Query: 297 MTSGNDTEDSDFKVDDQGVLRFRGKVCIPDNEELKKLILEESHKSRLSIHPGATKMYHDL 356
+ S N + D+ V V+ ++G++ +P++ +L K+I+ ESH S++ H G T+ +
Sbjct: 172 LCSNNAGKSGDY-VLHHDVIIWKGRIMLPNDSQLLKMIMTESHASKVGGHAGTTRTIVRI 348
Query: 357 KKLFWWSGLKRDVAQFVYACLICQKSKVEHQ-KPAGLL 393
F+W ++ D+ +FV C+ICQ++KV H PAGLL
Sbjct: 349 NAQFYWPKMREDIMKFVQECVICQQAKVTHSLLPAGLL 462
Score = 29.6 bits (65), Expect = 4.5
Identities = 13/21 (61%), Positives = 15/21 (70%)
Frame = +2
Query: 215 YDFGLNYHPG*ANVVADALSR 235
YDF + Y PG N+ ADALSR
Sbjct: 20 YDFIIQYSPGKENIPADALSR 82
>NP395548 reverse transcriptase [Glycine max]
Length = 762
Score = 54.3 bits (129), Expect = 2e-07
Identities = 28/63 (44%), Positives = 38/63 (59%)
Frame = +1
Query: 8 KCEFWLSEVSFLGHIIFGSDIAVDPSKVDAVSQWETLKSVTEIRSFLGLAGYYRRFIEGF 67
KC F + E LGH I I VD +K+D + + +V IRSFLG A +YRRFI+ F
Sbjct: 574 KCHFMVREGIVLGHKISTRGIEVDQTKIDVIEKLPPPSNVKGIRSFLGQARFYRRFIKDF 753
Query: 68 SKL 70
+K+
Sbjct: 754 TKV 762
>BE660092 weakly similar to GP|9884624|dbj retroelement pol polyprotein-like
{Arabidopsis thaliana}, partial (13%)
Length = 378
Score = 52.8 bits (125), Expect = 5e-07
Identities = 22/72 (30%), Positives = 42/72 (57%)
Frame = -3
Query: 492 RLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGGAWDSHLPLIEFTYNNSYHSSIGMAPF 551
R+S+ YHPQT+GQ+E + + ++ +L V W + L + + +Y + IGM+P+
Sbjct: 340 RVSTPYHPQTNGQAEISNREIKRILEKIVQPSRKDWSTRLDDALWAHRTAYKAPIGMSPY 161
Query: 552 EALYGRRCRTPL 563
++G+ C P+
Sbjct: 160 RVVFGKACHLPV 125
>BU549069
Length = 615
Score = 51.6 bits (122), Expect = 1e-06
Identities = 28/46 (60%), Positives = 32/46 (68%), Gaps = 7/46 (15%)
Frame = -1
Query: 621 LRVTPLTGVGRALKSRKLTPKFIGPYQISDR-------VGLPPHLS 659
L+VTP TGVG+ALKSRKLTP FIG +QI R + LPP LS
Sbjct: 615 LKVTPRTGVGQALKSRKLTPHFIGHFQILKRAXPVAYQIALPPSLS 478
>CA820403 weakly similar to GP|13273463|gb| pol protein integrase region
{Ginkgo biloba}, partial (52%)
Length = 421
Score = 50.8 bits (120), Expect = 2e-06
Identities = 30/76 (39%), Positives = 40/76 (52%), Gaps = 1/76 (1%)
Frame = -3
Query: 453 YIHSVVKLHGVPASIVSDRDPRFT-SRFWKRLQDALGSKLRLSSAYHPQTDGQSERTIQS 511
+I VKLHG +SIVSD D F S FW L G+KL+ S AYHPQ D ++ +
Sbjct: 335 FIKEAVKLHGCSSSIVSDWDRLFLIS*FWTELFKMEGTKLKFSLAYHPQPDSHTKVVNRC 156
Query: 512 LEDLLRVCVLEQGGAW 527
+E L+ + W
Sbjct: 155 IEMNLQCLTTSKRKQW 108
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.323 0.139 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 30,164,409
Number of Sequences: 63676
Number of extensions: 420885
Number of successful extensions: 1791
Number of sequences better than 10.0: 62
Number of HSP's better than 10.0 without gapping: 1766
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1776
length of query: 659
length of database: 12,639,632
effective HSP length: 103
effective length of query: 556
effective length of database: 6,081,004
effective search space: 3381038224
effective search space used: 3381038224
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 62 (28.5 bits)
Medicago: description of AC148291.4


