

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148289.7 - phase: 0
(197 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
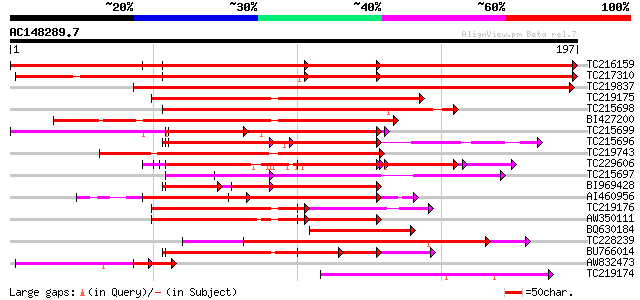
Score E
Sequences producing significant alignments: (bits) Value
TC216159 similar to UP|Q9SWF9 (Q9SWF9) Zinc finger protein, part... 365 e-102
TC217310 similar to UP|Q9SWF9 (Q9SWF9) Zinc finger protein, part... 339 4e-94
TC219837 similar to UP|Q9SWF9 (Q9SWF9) Zinc finger protein, part... 244 2e-65
TC219175 126 7e-30
TC215698 similar to UP|Q9FMJ7 (Q9FMJ7) Similarity to zinc finger... 125 1e-29
BI427200 124 2e-29
TC215699 similar to UP|Q9FMJ7 (Q9FMJ7) Similarity to zinc finger... 122 1e-28
TC215696 similar to UP|Q9FMJ7 (Q9FMJ7) Similarity to zinc finger... 120 3e-28
TC219743 similar to UP|Q94AD9 (Q94AD9) At1g04990/F13M7_1, partia... 115 2e-26
TC229606 similar to UP|Q941Q3 (Q941Q3) Floral homeotic protein H... 103 7e-23
TC215697 similar to UP|Q9FMJ7 (Q9FMJ7) Similarity to zinc finger... 92 1e-19
BI969428 80 3e-19
AI460956 90 6e-19
TC219176 similar to UP|Q6NPN3 (Q6NPN3) At5g18550, partial (11%) 86 1e-17
AW350111 82 1e-16
BQ630184 80 6e-16
TC228239 weakly similar to UP|Q6IVC4 (Q6IVC4) Zinc finger protei... 77 4e-15
BU766014 similar to GP|15081721|gb| At1g04990/F13M7_1 {Arabidops... 73 7e-14
AW832473 similar to PIR|T48868|T488 zinc finger protein [importe... 51 2e-09
TC219174 weakly similar to UP|Q6IVC4 (Q6IVC4) Zinc finger protei... 50 9e-07
>TC216159 similar to UP|Q9SWF9 (Q9SWF9) Zinc finger protein, partial (98%)
Length = 1871
Score = 365 bits (938), Expect = e-102
Identities = 170/197 (86%), Positives = 183/197 (92%)
Frame = +3
Query: 1 MGSDSPQQTMRNDQTYGTSHQGDPENAGLQGVYSQYRSGSVPVGFYALQRENIFPERPDQ 60
MGSDSPQQTM N Q+YGTS Q +P N+G QG YSQ+RSGSVPVGFYALQRENIFPERPDQ
Sbjct: 852 MGSDSPQQTMGNGQSYGTSRQSEPANSGSQGAYSQFRSGSVPVGFYALQRENIFPERPDQ 1031
Query: 61 PECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYSRYGICKF 120
PECQFYMKTGDCKFGAVCRFHHP ER IPAPDCVLSP+GLPLRPGEPLCVFYSRYGICKF
Sbjct: 1032 PECQFYMKTGDCKFGAVCRFHHPHERMIPAPDCVLSPIGLPLRPGEPLCVFYSRYGICKF 1211
Query: 121 GPSCKFDHPMGIFTYNVSASPLAEAAGRRLLGSSSGTAALSLSSEGLVESGSVKPRRLSL 180
GPSCKFDHPMG+FTYN+SASPLA+A GRR+LGSSSGT+AL+LSSEGLVESGS PRRLSL
Sbjct: 1212 GPSCKFDHPMGVFTYNMSASPLADAPGRRMLGSSSGTSALNLSSEGLVESGSANPRRLSL 1391
Query: 181 SETRPNPSGDDDIDDEG 197
SETR PSGDD+ID+EG
Sbjct: 1392 SETRQIPSGDDNIDEEG 1442
Score = 110 bits (275), Expect = 4e-25
Identities = 48/83 (57%), Positives = 58/83 (69%)
Frame = +3
Query: 47 ALQRENIFPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGE 106
A + + FPER QPECQ+Y+KTG CKFGA CRFHHPR++ A ++ LG PLRP E
Sbjct: 381 AARMKGEFPERIGQPECQYYLKTGTCKFGATCRFHHPRDKAGIAGRVAMNILGYPLRPNE 560
Query: 107 PLCVFYSRYGICKFGPSCKFDHP 129
P C +Y R G CKFG +CKF HP
Sbjct: 561 PECAYYLRTGQCKFGNTCKFHHP 629
Score = 82.0 bits (201), Expect = 2e-16
Identities = 33/76 (43%), Positives = 48/76 (62%)
Frame = +3
Query: 54 FPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYS 113
+PE P +P+C +Y++TG C+FGA CRF+HP R + + P R G+P C +Y
Sbjct: 267 YPEHPGEPDCSYYIRTGLCRFGATCRFNHPPNRKLAIAAARMKG-EFPERIGQPECQYYL 443
Query: 114 RYGICKFGPSCKFDHP 129
+ G CKFG +C+F HP
Sbjct: 444 KTGTCKFGATCRFHHP 491
Score = 56.6 bits (135), Expect = 7e-09
Identities = 23/51 (45%), Positives = 33/51 (64%)
Frame = +3
Query: 54 FPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRP 104
+P RP++PEC +Y++TG CKFG C+FHHP+ + VLS P+ P
Sbjct: 540 YPLRPNEPECAYYLRTGQCKFGNTCKFHHPQPNNM-----VLSMRSSPVYP 677
>TC217310 similar to UP|Q9SWF9 (Q9SWF9) Zinc finger protein, partial (93%)
Length = 1606
Score = 339 bits (870), Expect = 4e-94
Identities = 162/195 (83%), Positives = 174/195 (89%)
Frame = +1
Query: 3 SDSPQQTMRNDQTYGTSHQGDPENAGLQGVYSQYRSGSVPVGFYALQRENIFPERPDQPE 62
SDSPQQ MRN QTYGTS QG E AG QG YSQ+RSG+VPVGFY LQRENIFPERP QPE
Sbjct: 769 SDSPQQAMRNGQTYGTSRQG--ELAGSQGAYSQFRSGTVPVGFYTLQRENIFPERPGQPE 942
Query: 63 CQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYSRYGICKFGP 122
CQFYMKTGDCKFGAVCRFHHP+ER +PAP+CVLSP+G PLRPGEPLCVFYSRYGICKFGP
Sbjct: 943 CQFYMKTGDCKFGAVCRFHHPQERLVPAPNCVLSPIGPPLRPGEPLCVFYSRYGICKFGP 1122
Query: 123 SCKFDHPMGIFTYNVSASPLAEAAGRRLLGSSSGTAALSLSSEGLVESGSVKPRRLSLSE 182
SCKFDHPM IF++N+SASP A+A R LLGSSSGTAAL+LSSEGLVES S KPRRLSLSE
Sbjct: 1123SCKFDHPMEIFSHNISASPSADAPSRHLLGSSSGTAALNLSSEGLVESSSAKPRRLSLSE 1302
Query: 183 TRPNPSGDDDIDDEG 197
TR PSGDDDIDD+G
Sbjct: 1303TRQIPSGDDDIDDDG 1347
Score = 108 bits (269), Expect = 2e-24
Identities = 46/76 (60%), Positives = 55/76 (71%)
Frame = +1
Query: 54 FPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYS 113
FPER QPECQ+Y+KTG CKFGA C+FHHP+++ A L+ LG PLRP EP C +Y
Sbjct: 301 FPERIGQPECQYYLKTGTCKFGATCKFHHPKDQAGIAGRVALNILGYPLRPNEPECTYYL 480
Query: 114 RYGICKFGPSCKFDHP 129
R G CKFG +CKF HP
Sbjct: 481 RTGQCKFGNTCKFHHP 528
Score = 86.7 bits (213), Expect = 6e-18
Identities = 37/77 (48%), Positives = 51/77 (66%), Gaps = 1/77 (1%)
Frame = +1
Query: 54 FPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLG-LPLRPGEPLCVFY 112
+PERP +P+C +YM+TG C+FGA CRF+HP R + + +G P R G+P C +Y
Sbjct: 166 YPERPGEPDCSYYMRTGLCRFGATCRFNHPPNRKLAIATARM--IGEFPERIGQPECQYY 339
Query: 113 SRYGICKFGPSCKFDHP 129
+ G CKFG +CKF HP
Sbjct: 340 LKTGTCKFGATCKFHHP 390
Score = 57.0 bits (136), Expect = 6e-09
Identities = 23/51 (45%), Positives = 34/51 (66%)
Frame = +1
Query: 54 FPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRP 104
+P RP++PEC +Y++TG CKFG C+FHHP+ + +LS G P+ P
Sbjct: 439 YPLRPNEPECTYYLRTGQCKFGNTCKFHHPQPSNM-----MLSLRGSPVYP 576
>TC219837 similar to UP|Q9SWF9 (Q9SWF9) Zinc finger protein, partial (36%)
Length = 780
Score = 244 bits (623), Expect = 2e-65
Identities = 117/153 (76%), Positives = 128/153 (83%)
Frame = +2
Query: 44 GFYALQRENIFPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLR 103
GF + IFPER Q E FY+KTGDCKFGAVC+FHHPRER IPAPDCVLSP+GLPLR
Sbjct: 11 GFIHCRGRXIFPERXGQXEXXFYVKTGDCKFGAVCQFHHPRERLIPAPDCVLSPIGLPLR 190
Query: 104 PGEPLCVFYSRYGICKFGPSCKFDHPMGIFTYNVSASPLAEAAGRRLLGSSSGTAALSLS 163
GEPLCVFYSRYGICKFGPSCKFDHPM IF+YN++ SP A+A R LLGSSSGTAAL+LS
Sbjct: 191 LGEPLCVFYSRYGICKFGPSCKFDHPMEIFSYNITTSPSADAPSRHLLGSSSGTAALNLS 370
Query: 164 SEGLVESGSVKPRRLSLSETRPNPSGDDDIDDE 196
SEGLVES S KPR LSLSE R PSGDD+IDD+
Sbjct: 371 SEGLVESSSAKPRPLSLSEIRQIPSGDDNIDDD 469
>TC219175
Length = 561
Score = 126 bits (316), Expect = 7e-30
Identities = 52/95 (54%), Positives = 70/95 (72%)
Frame = +2
Query: 50 RENIFPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLC 109
+E ++PERP +PECQ+Y++TGDCKFG C++HHP + + P +LSP+GLPLRPG C
Sbjct: 107 KEQLYPERPGEPECQYYLRTGDCKFGLACQYHHPHDHIVAQP--LLSPVGLPLRPGLQPC 280
Query: 110 VFYSRYGICKFGPSCKFDHPMGIFTYNVSASPLAE 144
FY + G CKFG +CKFDHP+G Y+ SAS L +
Sbjct: 281 AFYLQNGHCKFGSTCKFDHPLGSMRYSPSASSLID 385
>TC215698 similar to UP|Q9FMJ7 (Q9FMJ7) Similarity to zinc finger protein,
partial (17%)
Length = 727
Score = 125 bits (314), Expect = 1e-29
Identities = 59/111 (53%), Positives = 73/111 (65%), Gaps = 8/111 (7%)
Frame = +1
Query: 54 FPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYS 113
FPERP +PEC F++KTGDCKF + C+FHHP+ R P C LS GLPLRP + +C YS
Sbjct: 130 FPERPGEPECSFFLKTGDCKFKSNCKFHHPKNRVTRLPPCNLSDKGLPLRPDQSVCSHYS 309
Query: 114 RYGICKFGPSCKFDHPM--------GIFTYNVSASPLAEAAGRRLLGSSSG 156
RYGICKFGP+CKFDHP+ G+ + S S A AG +G S+G
Sbjct: 310 RYGICKFGPACKFDHPINLQPVMIPGLEQQSYSNSASAGVAG---IGGSTG 453
>BI427200
Length = 421
Score = 124 bits (312), Expect = 2e-29
Identities = 58/120 (48%), Positives = 78/120 (64%)
Frame = +3
Query: 16 YGTSHQGDPENAGLQGVYSQYRSGSVPVGFYALQRENIFPERPDQPECQFYMKTGDCKFG 75
YG + P +A + Y+ S + P G + ++ FPERP +PECQ+Y++TGDCKFG
Sbjct: 6 YGVTQLSSPTSAFARP-YTPLSSTTGPSG--SNLKDQFFPERPGEPECQYYLRTGDCKFG 176
Query: 76 AVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYSRYGICKFGPSCKFDHPMGIFTY 135
CR+HHPR+ + P +LSP+GLPLRPG C FY + G CKFG +CKFDHP+G Y
Sbjct: 177 LACRYHHPRDHIVARP--LLSPVGLPLRPGVQPCAFYLQNGHCKFGSTCKFDHPLGSTRY 350
>TC215699 similar to UP|Q9FMJ7 (Q9FMJ7) Similarity to zinc finger protein,
partial (34%)
Length = 1124
Score = 122 bits (306), Expect = 1e-28
Identities = 62/147 (42%), Positives = 80/147 (54%), Gaps = 15/147 (10%)
Frame = +1
Query: 1 MGSDSPQQTMRNDQTYGTSHQGDPENAGLQGVYSQYRSGSVPVGF-----------YALQ 49
+ SP M T G S Q N VY R+ P F Y
Sbjct: 349 LNESSPFVPMMLSPTQGVSTQSSDWNGYQASVYLPERNMHPPSTFVMNNPAIDTNVYMHH 528
Query: 50 RENI----FPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPG 105
++ + FPERP +PEC +++KTGDCKF + C+F+HP+ R P C LS GLPLRP
Sbjct: 529 QKQMPVDEFPERPGEPECSYFLKTGDCKFKSNCKFNHPKNRVARLPPCNLSDKGLPLRPD 708
Query: 106 EPLCVFYSRYGICKFGPSCKFDHPMGI 132
+ +C YSRYGICKFGP+CKFDHP+ +
Sbjct: 709 QSVCSHYSRYGICKFGPACKFDHPINL 789
Score = 89.0 bits (219), Expect = 1e-18
Identities = 39/76 (51%), Positives = 52/76 (68%), Gaps = 2/76 (2%)
Frame = +1
Query: 56 ERPDQPECQFYMKTGDCKFGAVCRFHHPRER--TIPAPDCVLSPLGLPLRPGEPLCVFYS 113
ER EC++Y ++G CKFG C+++H R + T PAP L+ LGLP+RPGE C +Y
Sbjct: 1 ERSGMTECKYYQRSGGCKFGKSCKYNHTRGKISTAPAPLLELNFLGLPIRPGERECPYYM 180
Query: 114 RYGICKFGPSCKFDHP 129
R G CKFG +CKF+HP
Sbjct: 181 RTGSCKFGANCKFNHP 228
Score = 49.3 bits (116), Expect = 1e-06
Identities = 18/29 (62%), Positives = 23/29 (79%)
Frame = +1
Query: 55 PERPDQPECQFYMKTGDCKFGAVCRFHHP 83
P RP + EC +YM+TG CKFGA C+F+HP
Sbjct: 142 PIRPGERECPYYMRTGSCKFGANCKFNHP 228
Score = 38.1 bits (87), Expect = 0.003
Identities = 18/42 (42%), Positives = 24/42 (56%)
Frame = +1
Query: 103 RPGEPLCVFYSRYGICKFGPSCKFDHPMGIFTYNVSASPLAE 144
R G C +Y R G CKFG SCK++H G + + +PL E
Sbjct: 4 RSGMTECKYYQRSGGCKFGKSCKYNHTRG--KISTAPAPLLE 123
>TC215696 similar to UP|Q9FMJ7 (Q9FMJ7) Similarity to zinc finger protein,
partial (45%)
Length = 1204
Score = 120 bits (302), Expect = 3e-28
Identities = 48/76 (63%), Positives = 59/76 (77%)
Frame = +1
Query: 54 FPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYS 113
FPERP +PEC +++KTGDCKF + C+FHHP+ R P C LS GLPLRP + +C +Y
Sbjct: 673 FPERPGEPECSYFLKTGDCKFKSNCKFHHPKNRIARLPLCNLSDKGLPLRPDQNVCTYYR 852
Query: 114 RYGICKFGPSCKFDHP 129
RYGICKFGP+CKFDHP
Sbjct: 853 RYGICKFGPACKFDHP 900
Score = 85.9 bits (211), Expect = 1e-17
Identities = 50/131 (38%), Positives = 71/131 (54%), Gaps = 1/131 (0%)
Frame = +1
Query: 56 ERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCV-LSPLGLPLRPGEPLCVFYSR 114
ER Q EC++Y+++G CKFG C+F+H R ++ A L+ LGLP+R GE C +Y R
Sbjct: 130 ERSGQTECKYYLRSGGCKFGKACKFNHTRGKSSSASSTAELNFLGLPIRVGEKECHYYMR 309
Query: 115 YGICKFGPSCKFDHPMGIFTYNVSASPLAEAAGRRLLGSSSGTAALSLSSEGLVESGSVK 174
G CKFG +C+F+HP P A G G +G+ S+S +G+ +S
Sbjct: 310 TGSCKFGANCRFNHP----------DPTAIGGGDSPSGYGNGS---SISLQGVSQSSI-- 444
Query: 175 PRRLSLSETRP 185
S S TRP
Sbjct: 445 ---SSWSSTRP 468
Score = 48.9 bits (115), Expect = 1e-06
Identities = 23/45 (51%), Positives = 27/45 (59%)
Frame = +1
Query: 55 PERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLG 99
P R + EC +YM+TG CKFGA CRF+HP I D SP G
Sbjct: 268 PIRVGEKECHYYMRTGSCKFGANCRFNHPDPTAIGGGD---SPSG 393
Score = 45.1 bits (105), Expect = 2e-05
Identities = 18/38 (47%), Positives = 22/38 (57%)
Frame = +1
Query: 55 PERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPD 92
P RPDQ C +Y + G CKFG C+F HP T+ D
Sbjct: 814 PLRPDQNVCTYYRRYGICKFGPACKFDHPAPSTMAGHD 927
>TC219743 similar to UP|Q94AD9 (Q94AD9) At1g04990/F13M7_1, partial (17%)
Length = 733
Score = 115 bits (287), Expect = 2e-26
Identities = 49/99 (49%), Positives = 69/99 (69%)
Frame = +2
Query: 32 VYSQYRSGSVPVGFYALQRENIFPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAP 91
VY G G A+ + P RPDQPEC+++M TG CK+G+ C+FHHP+ER +
Sbjct: 116 VYDYMNLGESLFGGQAIN--SALPNRPDQPECRYFMSTGTCKYGSDCKFHHPKERMSQS- 286
Query: 92 DCVLSPLGLPLRPGEPLCVFYSRYGICKFGPSCKFDHPM 130
+++PLGLP+RPG+ +C +Y YG+CKFGP+CKFDHP+
Sbjct: 287 --LINPLGLPVRPGQAVCSYYRIYGMCKFGPTCKFDHPV 397
>TC229606 similar to UP|Q941Q3 (Q941Q3) Floral homeotic protein HUA1, partial
(50%)
Length = 1569
Score = 103 bits (256), Expect = 7e-23
Identities = 49/113 (43%), Positives = 66/113 (58%), Gaps = 4/113 (3%)
Frame = +3
Query: 51 ENIFPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPA----PDCVLSPLGLPLRPGE 106
+ I+P+RP Q EC FYMKTG+CKFG C++HHP +R+ P+ L+P GLP R G
Sbjct: 1005 DTIYPQRPGQIECDFYMKTGECKFGERCKYHHPIDRSAPSLSKQATVKLTPAGLPRREGA 1184
Query: 107 PLCVFYSRYGICKFGPSCKFDHPMGIFTYNVSASPLAEAAGRRLLGSSSGTAA 159
+C +Y + G CKFG +CKFDHP ++ S A G G T+A
Sbjct: 1185 VICPYYLKTGTCKFGATCKFDHPPPGEVMEMAKSQGTSANGGEAEGDEIETSA 1343
Score = 87.8 bits (216), Expect = 3e-18
Identities = 41/104 (39%), Positives = 66/104 (63%), Gaps = 2/104 (1%)
Frame = +3
Query: 55 PERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYSR 114
PERP +P+C +++KT CKFG+ C+F+HP+ + + + +S GLP RP EP C FY +
Sbjct: 402 PERPGEPDCPYFLKTQRCKFGSKCKFNHPK---VSSENADVSS-GLPERPSEPPCAFYMK 569
Query: 115 YGICKFGPSCKFDHP--MGIFTYNVSASPLAEAAGRRLLGSSSG 156
G C++G +CKF HP + I N S+ +A+ ++G ++G
Sbjct: 570 TGKCRYGAACKFHHPKDIQIQLSNDSSQTVAQTQTNSIMGWATG 701
Score = 83.6 bits (205), Expect = 5e-17
Identities = 49/153 (32%), Positives = 72/153 (47%), Gaps = 31/153 (20%)
Frame = +3
Query: 55 PERPDQPECQFYMKTGDCKFGAVCRFHHPRERTI-------------------------- 88
PERP +P C FYMKTG C++GA C+FHHP++ I
Sbjct: 528 PERPSEPPCAFYMKTGKCRYGAACKFHHPKDIQIQLSNDSSQTVAQTQTNSIMGWATGDT 707
Query: 89 PAPDCVLSPL-----GLPLRPGEPLCVFYSRYGICKFGPSCKFDHPMGIFTYNVSASPLA 143
P ++SP GLP+R GE C FY + G CK+G +C+++HP N P+A
Sbjct: 708 PPIQSLISPSLQNSKGLPVRLGEVDCPFYMKTGSCKYGVTCRYNHP----DRNAINPPIA 875
Query: 144 EAAGRRLLGSSSGTAALSLSSEGLVESGSVKPR 176
G + SS+ + L + + + +PR
Sbjct: 876 -GLGASIFPSSAANLNIGLLNPAVSVYQAFEPR 971
Score = 70.1 bits (170), Expect = 6e-13
Identities = 35/82 (42%), Positives = 47/82 (56%), Gaps = 5/82 (6%)
Frame = +3
Query: 53 IFPERPDQPECQFYMKTGDCKFGAVCRFHHP---RERTIP--APDCVLSPLGLPLRPGEP 107
I+P+RP + +C YM T CKFG C+F HP E IP +++ P RPGEP
Sbjct: 243 IYPQRPGEKDCAHYMLTRTCKFGDSCKFDHPFWVPEGGIPDWKEVPIVTSETPPERPGEP 422
Query: 108 LCVFYSRYGICKFGPSCKFDHP 129
C ++ + CKFG CKF+HP
Sbjct: 423 DCPYFLKTQRCKFGSKCKFNHP 488
Score = 67.0 bits (162), Expect = 5e-12
Identities = 39/124 (31%), Positives = 51/124 (40%), Gaps = 40/124 (32%)
Frame = +3
Query: 47 ALQRENIFPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVL----------- 95
+LQ P R + +C FYMKTG CK+G CR++HP I P L
Sbjct: 735 SLQNSKGLPVRLGEVDCPFYMKTGSCKYGVTCRYNHPDRNAINPPIAGLGASIFPSSAAN 914
Query: 96 ------------------------SPLGL-----PLRPGEPLCVFYSRYGICKFGPSCKF 126
S +G+ P RPG+ C FY + G CKFG CK+
Sbjct: 915 LNIGLLNPAVSVYQAFEPRLSNPMSQVGIADTIYPQRPGQIECDFYMKTGECKFGERCKY 1094
Query: 127 DHPM 130
HP+
Sbjct: 1095HHPI 1106
Score = 43.5 bits (101), Expect = 6e-05
Identities = 18/32 (56%), Positives = 19/32 (59%)
Frame = +3
Query: 101 PLRPGEPLCVFYSRYGICKFGPSCKFDHPMGI 132
P RPGE C Y CKFG SCKFDHP +
Sbjct: 249 PQRPGEKDCAHYMLTRTCKFGDSCKFDHPFWV 344
>TC215697 similar to UP|Q9FMJ7 (Q9FMJ7) Similarity to zinc finger protein,
partial (12%)
Length = 798
Score = 92.4 bits (228), Expect = 1e-19
Identities = 43/101 (42%), Positives = 58/101 (56%)
Frame = +2
Query: 72 CKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYSRYGICKFGPSCKFDHPMG 131
CKF + C+FHHP+ R P C LS GLPLRP + +C +Y RYGICKFGP+CKFDHP
Sbjct: 287 CKFKSNCKFHHPKNRIARLPPCNLSDKGLPLRPEQTVCTYYRRYGICKFGPACKFDHP-- 460
Query: 132 IFTYNVSASPLAEAAGRRLLGSSSGTAALSLSSEGLVESGS 172
P + AG S + +A + ++ G + G+
Sbjct: 461 ---------PPSTMAGLDQQSSYTNSAGVDVAENGDADDGN 556
Score = 43.1 bits (100), Expect = 8e-05
Identities = 17/38 (44%), Positives = 22/38 (57%)
Frame = +2
Query: 55 PERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPD 92
P RP+Q C +Y + G CKFG C+F HP T+ D
Sbjct: 374 PLRPEQTVCTYYRRYGICKFGPACKFDHPPPSTMAGLD 487
>BI969428
Length = 536
Score = 79.7 bits (195), Expect(2) = 3e-19
Identities = 32/52 (61%), Positives = 38/52 (72%)
Frame = -3
Query: 78 CRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYSRYGICKFGPSCKFDHP 129
C+FHHP+ R P C LS GLPL P + +C +Y RYGICKFGP+CKFDHP
Sbjct: 411 CKFHHPKNRIARLPLCNLSGKGLPLSPDQNVCPYYRRYGICKFGPACKFDHP 256
Score = 40.8 bits (94), Expect = 4e-04
Identities = 16/38 (42%), Positives = 20/38 (52%)
Frame = -3
Query: 55 PERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPD 92
P PDQ C +Y + G CKFG C+F HP + D
Sbjct: 342 PLSPDQNVCPYYRRYGICKFGPACKFDHPAPSAMAGHD 229
Score = 32.0 bits (71), Expect(2) = 3e-19
Identities = 11/21 (52%), Positives = 15/21 (71%)
Frame = -2
Query: 54 FPERPDQPECQFYMKTGDCKF 74
FPER +PEC +++ TG KF
Sbjct: 484 FPERSGEPECSYFLNTGXXKF 422
>AI460956
Length = 315
Score = 90.1 bits (222), Expect = 6e-19
Identities = 40/83 (48%), Positives = 51/83 (61%)
Frame = +2
Query: 47 ALQRENIFPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGE 106
A++ +PER +P CQ+Y+KTG CKFGA C+FHHP+ L+ G PLR GE
Sbjct: 56 AVRATGDYPERVGEPPCQYYLKTGTCKFGASCKFHHPKNGGGYLSQAPLNIYGYPLRLGE 235
Query: 107 PLCVFYSRYGICKFGPSCKFDHP 129
C +Y + CKFG SCKF HP
Sbjct: 236 KECSYYXKTXQCKFGISCKFHHP 304
Score = 59.3 bits (142), Expect = 1e-09
Identities = 32/66 (48%), Positives = 39/66 (58%)
Frame = +2
Query: 77 VCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYSRYGICKFGPSCKFDHPMGIFTYN 136
VCRF+HPR+R A V + P R GEP C +Y + G CKFG SCKF HP Y
Sbjct: 11 VCRFNHPRDRAAVAA-AVRATGDYPERVGEPPCQYYLKTGTCKFGASCKFHHPKNGGGY- 184
Query: 137 VSASPL 142
+S +PL
Sbjct: 185 LSQAPL 202
Score = 45.8 bits (107), Expect = 1e-05
Identities = 24/61 (39%), Positives = 31/61 (50%)
Frame = +2
Query: 24 PENAGLQGVYSQYRSGSVPVGFYALQRENIFPERPDQPECQFYMKTGDCKFGAVCRFHHP 83
P+N G G SQ P+ Y +P R + EC +Y KT CKFG C+FHHP
Sbjct: 164 PKNGG--GYLSQ-----APLNIYG------YPLRLGEKECSYYXKTXQCKFGISCKFHHP 304
Query: 84 R 84
+
Sbjct: 305 Q 307
>TC219176 similar to UP|Q6NPN3 (Q6NPN3) At5g18550, partial (11%)
Length = 539
Score = 85.5 bits (210), Expect = 1e-17
Identities = 33/55 (60%), Positives = 45/55 (81%)
Frame = +1
Query: 50 RENIFPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRP 104
+E ++PERP +PECQ+Y++TGDCKFG CR+HHPR+ + P +LSP+GLPLRP
Sbjct: 298 KEQLYPERPGEPECQYYLRTGDCKFGLACRYHHPRDHIVARP--LLSPVGLPLRP 456
Score = 45.8 bits (107), Expect = 1e-05
Identities = 21/47 (44%), Positives = 27/47 (56%)
Frame = +1
Query: 101 PLRPGEPLCVFYSRYGICKFGPSCKFDHPMGIFTYNVSASPLAEAAG 147
P RPGEP C +Y R G CKFG +C++ HP ++ A PL G
Sbjct: 313 PERPGEPECQYYLRTGDCKFGLACRYHHPRD----HIVARPLLSPVG 441
>AW350111
Length = 641
Score = 82.4 bits (202), Expect = 1e-16
Identities = 35/55 (63%), Positives = 45/55 (81%)
Frame = -1
Query: 50 RENIFPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRP 104
++ FPERP +PECQ+Y++TGDCKFG CR+HHPR+ I AP +LSP+GLPLRP
Sbjct: 542 KDRFFPERPGEPECQYYLRTGDCKFGLACRYHHPRDH-IVAP--LLSPVGLPLRP 387
Score = 44.7 bits (104), Expect = 3e-05
Identities = 17/29 (58%), Positives = 21/29 (71%)
Frame = -1
Query: 101 PLRPGEPLCVFYSRYGICKFGPSCKFDHP 129
P RPGEP C +Y R G CKFG +C++ HP
Sbjct: 527 PERPGEPECQYYLRTGDCKFGLACRYHHP 441
>BQ630184
Length = 429
Score = 80.1 bits (196), Expect = 6e-16
Identities = 33/37 (89%), Positives = 36/37 (97%)
Frame = +1
Query: 105 GEPLCVFYSRYGICKFGPSCKFDHPMGIFTYNVSASP 141
GEPLCVFYSRYGICKFGPSCKFDHPM IF++N+SASP
Sbjct: 319 GEPLCVFYSRYGICKFGPSCKFDHPMEIFSHNISASP 429
Score = 37.7 bits (86), Expect = 0.003
Identities = 14/26 (53%), Positives = 17/26 (64%)
Frame = +1
Query: 60 QPECQFYMKTGDCKFGAVCRFHHPRE 85
+P C FY + G CKFG C+F HP E
Sbjct: 322 EPLCVFYSRYGICKFGPSCKFDHPME 399
>TC228239 weakly similar to UP|Q6IVC4 (Q6IVC4) Zinc finger protein
(Zinc-finger transcription factor), partial (32%)
Length = 1244
Score = 77.4 bits (189), Expect = 4e-15
Identities = 41/87 (47%), Positives = 53/87 (60%), Gaps = 1/87 (1%)
Frame = +2
Query: 82 HPRERTIPAPDCVLSPLGLPLRPGEPLCVFYSRYGICKFGPSCKFDHPMGIFTYNVSASP 141
HP +++ P LSP+GL R P C Y++ G+CKFG +CKFDHPMG +Y+ SAS
Sbjct: 2 HPPDKSAPIATVTLSPVGLS*RTVAPPCTHYTQRGVCKFGSACKFDHPMGSLSYSPSASS 181
Query: 142 LAE-AAGRRLLGSSSGTAALSLSSEGL 167
LA+ +GSS GT A S SS L
Sbjct: 182 LADMPVAPYPVGSSIGTLAPSSSSSEL 262
Score = 42.0 bits (97), Expect = 2e-04
Identities = 33/121 (27%), Positives = 51/121 (41%)
Frame = +2
Query: 61 PECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYSRYGICKF 120
P C Y + G CKFG+ C+F HP +P S L P P V S +
Sbjct: 77 PPCTHYTQRGVCKFGSACKFDHPMGSLSYSP----SASSLADMPVAPYPVGSSIGTLAPS 244
Query: 121 GPSCKFDHPMGIFTYNVSASPLAEAAGRRLLGSSSGTAALSLSSEGLVESGSVKPRRLSL 180
S + +G ++ + A+ + +S+G+ L+LSS G + S +P S
Sbjct: 245 SSSSELRPELG-----AGSNKESVASRMSSMSTSTGSVGLTLSSVGPISHSSTQPSAQSS 409
Query: 181 S 181
S
Sbjct: 410 S 412
>BU766014 similar to GP|15081721|gb| At1g04990/F13M7_1 {Arabidopsis
thaliana}, partial (16%)
Length = 409
Score = 73.2 bits (178), Expect = 7e-14
Identities = 30/76 (39%), Positives = 48/76 (62%)
Frame = +3
Query: 54 FPERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYS 113
+P+RP +PEC +Y++TG +G+ CR+HHP +I LP R G+P C ++
Sbjct: 96 YPDRPGEPECLYYLRTGAFFYGSNCRYHHPAHISIGTH----YGEELPQRAGQPDCEYFL 263
Query: 114 RYGICKFGPSCKFDHP 129
+ G+CK+G +CK+ HP
Sbjct: 264 KTGMCKYGSTCKYHHP 311
Score = 68.6 bits (166), Expect = 2e-12
Identities = 27/62 (43%), Positives = 42/62 (67%)
Frame = +3
Query: 55 PERPDQPECQFYMKTGDCKFGAVCRFHHPRERTIPAPDCVLSPLGLPLRPGEPLCVFYSR 114
P+R QP+C++++KTG CK+G+ C++HHP++R AP + LG P+R E C +Y R
Sbjct: 225 PQRAGQPDCEYFLKTGMCKYGSTCKYHHPKDRRGAAP-VSFNTLGFPMRQEEKSCPYYMR 401
Query: 115 YG 116
G
Sbjct: 402 TG 407
Score = 40.0 bits (92), Expect = 7e-04
Identities = 18/48 (37%), Positives = 26/48 (53%)
Frame = +3
Query: 101 PLRPGEPLCVFYSRYGICKFGPSCKFDHPMGIFTYNVSASPLAEAAGR 148
P RPGEP C++Y R G +G +C++ HP I L + AG+
Sbjct: 99 PDRPGEPECLYYLRTGAFFYGSNCRYHHPAHISIGTHYGEELPQRAGQ 242
Score = 30.4 bits (67), Expect = 0.55
Identities = 14/34 (41%), Positives = 19/34 (55%)
Frame = +3
Query: 37 RSGSVPVGFYALQRENIFPERPDQPECQFYMKTG 70
R G+ PV F L FP R ++ C +YM+TG
Sbjct: 318 RRGAAPVSFNTLG----FPMRQEEKSCPYYMRTG 407
>AW832473 similar to PIR|T48868|T488 zinc finger protein [imported] - garden
pea, partial (14%)
Length = 319
Score = 51.2 bits (121), Expect(2) = 2e-09
Identities = 29/64 (45%), Positives = 34/64 (52%), Gaps = 16/64 (25%)
Frame = +3
Query: 3 SDSPQQTMRNDQTYGTSHQGDPENAGLQG----------------VYSQYRSGSVPVGFY 46
SDSPQQ MRN QTYGTS QG+ Q ++RSG+VPVGFY
Sbjct: 102 SDSPQQAMRNGQTYGTSRQGELAGXPTQKKGGXAPNFXNFXXPPFXPQKFRSGTVPVGFY 281
Query: 47 ALQR 50
LQ+
Sbjct: 282 TLQK 293
Score = 27.3 bits (59), Expect(2) = 2e-09
Identities = 11/15 (73%), Positives = 12/15 (79%)
Frame = +1
Query: 44 GFYALQRENIFPERP 58
GF +RENIFPERP
Sbjct: 274 GFIHCRRENIFPERP 318
>TC219174 weakly similar to UP|Q6IVC4 (Q6IVC4) Zinc finger protein
(Zinc-finger transcription factor), partial (31%)
Length = 710
Score = 49.7 bits (117), Expect = 9e-07
Identities = 33/85 (38%), Positives = 43/85 (49%), Gaps = 4/85 (4%)
Frame = +1
Query: 109 CVFYSRYGICKFGPSCKFDHPMGIFTYNVSASPLAEAAGRRL-LGSSSGTAALSLSSEGL 167
C FY + G CKFG +CKFDHP+G Y+ SAS L + +GS A S +S L
Sbjct: 7 CAFYLQNGHCKFGSTCKFDHPLGSMRYSPSASSLIDVPVTPYPVGSLLSQLAPSTTSSDL 186
Query: 168 ---VESGSVKPRRLSLSETRPNPSG 189
+ SGS K + + N SG
Sbjct: 187 RPELMSGSKKESFSARIPSSGNSSG 261
Score = 38.9 bits (89), Expect = 0.002
Identities = 31/122 (25%), Positives = 47/122 (38%), Gaps = 5/122 (4%)
Frame = +1
Query: 63 CQFYMKTGDCKFGAVCRFHHP--RERTIPAPDCVLSPLGLPLRPGEPLCVFYSRYGICKF 120
C FY++ G CKFG+ C+F HP R P+ ++ P G L
Sbjct: 7 CAFYLQNGHCKFGSTCKFDHPLGSMRYSPSASSLIDVPVTPYPVGSLLSQLAPSTTSSDL 186
Query: 121 GPSCKFDHPMGIFTYNVSASPLAEAAGRRLLGSSSGTAALS---LSSEGLVESGSVKPRR 177
P F+ + +S + L+ S G+ +LS LSS+ S + R
Sbjct: 187 RPELMSGSKKESFSARIPSSGNSSGTSVGLIFSQGGSVSLSDVQLSSQSNAPPNSSRSTR 366
Query: 178 LS 179
S
Sbjct: 367 QS 372
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.137 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,203,119
Number of Sequences: 63676
Number of extensions: 193427
Number of successful extensions: 1023
Number of sequences better than 10.0: 60
Number of HSP's better than 10.0 without gapping: 910
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 978
length of query: 197
length of database: 12,639,632
effective HSP length: 92
effective length of query: 105
effective length of database: 6,781,440
effective search space: 712051200
effective search space used: 712051200
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 56 (26.2 bits)
Medicago: description of AC148289.7


