

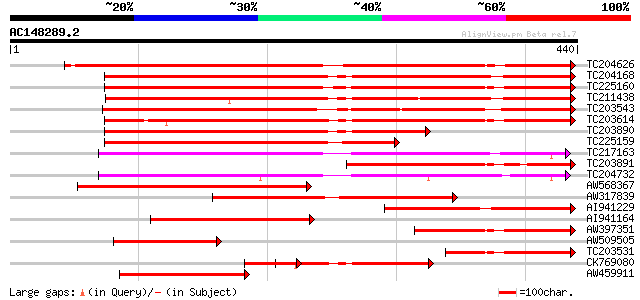
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148289.2 + phase: 0 /pseudo
(440 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204626 UP|GLCB_SOYBN (P25974) Beta-conglycinin, beta chain pre... 426 e-120
TC204168 UP|Q7XXT2 (Q7XXT2) Prepro beta-conglycinin alpha prime ... 407 e-114
TC225160 UP|Q948X9 (Q948X9) Beta-conglycinin alpha-subunit, comp... 400 e-112
TC211438 UP|GLCX_SOYBN (P11827) Beta-conglycinin, alpha' chain p... 382 e-106
TC203543 similar to UP|O22121 (O22121) Beta subunit of beta cong... 381 e-106
TC203614 UP|Q94LX2 (Q94LX2) Beta-conglycinin alpha subunit, comp... 380 e-106
TC203890 homologue to UP|Q7XXT2 (Q7XXT2) Prepro beta-conglycinin... 228 3e-60
TC225159 homologue to UP|O22120 (O22120) Alpha subunit of beta c... 209 2e-54
TC217163 UP|Q84V19 (Q84V19) Sucrose-binding protein 2, complete 199 2e-51
TC203891 homologue to UP|GLCA_SOYBN (P13916) Beta-conglycinin, a... 198 4e-51
TC204732 homologue to UP|SBP_SOYBN (Q04672) Sucrose-binding prot... 189 2e-48
AW568367 similar to GP|15425635|db beta-conglycinin beta-subunit... 180 1e-45
AW317839 similar to PIR|S20007|S20 beta-conglycinin alpha chain ... 159 2e-39
AI941229 similar to PIR|S20007|S20 beta-conglycinin alpha chain ... 151 5e-37
AI941164 similar to PIR|S20007|S20 beta-conglycinin alpha chain ... 120 2e-27
AW397351 similar to PIR|S20007|S20 beta-conglycinin alpha chain ... 117 1e-26
AW509505 114 1e-25
TC203531 UP|GLCA_SOYBN (P13916) Beta-conglycinin, alpha chain pr... 110 1e-24
CK769080 109 3e-24
AW459911 similar to GP|15425631|db beta-conglycinin alpha prime ... 108 5e-24
>TC204626 UP|GLCB_SOYBN (P25974) Beta-conglycinin, beta chain precursor,
complete
Length = 1653
Score = 426 bits (1096), Expect = e-120
Identities = 221/398 (55%), Positives = 295/398 (73%), Gaps = 1/398 (0%)
Frame = +2
Query: 43 MAIKAPCSLLMLLGIVFLASICVSSRSDQDQENPFIF-NSNRFQTLFENENGHIRLLQRF 101
M ++ P LL+LLG VFLAS+CVS + +D+ NPF +SN FQTLFEN+NG IRLLQRF
Sbjct: 23 MRVRFP--LLVLLGTVFLASVCVSLKVREDENNPFYLRSSNSFQTLFENQNGRIRLLQRF 196
Query: 102 DKRSKIFENLQNYRLLEYHSKPHTLFLPQHNDADFILVVVSGKAILTVLNPNNRNSFNLE 161
+KRS ENL++YR++++ SKP+T+ LP H DADF+L V+SG+AILT++N ++R+S+NL
Sbjct: 197 NKRSPQLENLRDYRIVQFQSKPNTILLPHHADADFLLFVLSGRAILTLVNNDDRDSYNLH 376
Query: 162 RGDTIKLPAGTLGYLANRDDNKDLRVLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKN 221
GD ++PAGT YL N D+++L+++ LAIPVN+PG++ F LS ++ QQS+L GFS N
Sbjct: 377 PGDAQRIPAGTTYYLVNPHDHQNLKIIKLAIPVNKPGRYDDFFLSSTQAQQSYLQGFSHN 556
Query: 222 ILEAAFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQEANVIVKVSREQIEELSKN 281
ILE +F+S +EEI RVL+ E E +Q Q+ VIV++S+EQI +LS+
Sbjct: 557 ILETSFHSEFEEINRVLLGEEE---------------EQRQQEGVIVELSKEQIRQLSRR 691
Query: 282 AKSSSRRSESSESEPINLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIREGSL 341
AKSSSR++ SSE EP NLR++ PIYSN FG FFEITPEKNPQL+DLDI ++ +I EG+L
Sbjct: 692 AKSSSRKTISSEDEPFNLRSRNPIYSNNFGKFFEITPEKNPQLRDLDIFLSSVDINEGAL 871
Query: 342 LLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQQQERSQQVQRYRARLSP 401
LLPHFNS+A VI+ + EG ELVG + E QQ+Q+ Q+E +VQRYRA LS
Sbjct: 872 LLPHFNSKAIVILVINEGDANIELVGIK-EQQQKQK-------QEEEPLEVQRYRAELSE 1027
Query: 402 GDVYVIPAGHPIVVTASSDLSLLGFGINAENNQRNFLA 439
DV+VIPA +P VV A+S+L+ L FGINAENNQRNFLA
Sbjct: 1028DDVFVIPAAYPFVVNATSNLNFLAFGINAENNQRNFLA 1141
Score = 32.7 bits (73), Expect = 0.33
Identities = 12/17 (70%), Positives = 16/17 (93%)
Frame = +1
Query: 74 ENPFIFNSNRFQTLFEN 90
+NPF+F SNRF+TLF+N
Sbjct: 1603 KNPFLFGSNRFETLFKN 1653
>TC204168 UP|Q7XXT2 (Q7XXT2) Prepro beta-conglycinin alpha prime subunit,
complete
Length = 2055
Score = 407 bits (1047), Expect = e-114
Identities = 197/366 (53%), Positives = 286/366 (77%)
Frame = +2
Query: 74 ENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHND 133
+NPF FNS RFQTLF+N+ GH+R+LQRF+KRS+ +NL++YR+LE++SKP+TL LP H D
Sbjct: 683 KNPFHFNSKRFQTLFKNQYGHVRVLQRFNKRSQQLQNLRDYRILEFNSKPNTLLLPHHAD 862
Query: 134 ADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDLRVLDLAIP 193
AD+++V+++G AILT++N ++R+S+NL+ GD +++PAGT Y+ N D++++LR++ LAIP
Sbjct: 863 ADYLIVILNGTAILTLVNNDDRDSYNLQSGDALRVPAGTTYYVVNPDNDENLRMITLAIP 1042
Query: 194 VNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEPQHRRGLR 253
VN+PG+F+SF LS ++ QQS+L GFSKNILEA++++ +EEI +VL E + Q
Sbjct: 1043 VNKPGRFESFFLSSTQAQQSYLQGFSKNILEASYDTKFEEINKVLFGREEGQQQ------ 1204
Query: 254 KDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQKPIYSNKFGNF 313
+ER Q+S VIV++S++QI ELSK+AKSSSR++ SSE +P NLR++ PIYSNK G
Sbjct: 1205 GEERLQES----VIVEISKKQIRELSKHAKSSSRKTISSEDKPFNLRSRDPIYSNKLGKL 1372
Query: 314 FEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQ 373
FEITPEKNPQL+DLD+ ++ ++ EG+L LPHFNS+A V++ + EG+ ELVG + + Q
Sbjct: 1373 FEITPEKNPQLRDLDVFLSVVDMNEGALFLPHFNSKAIVVLVINEGEANIELVGIKEQQQ 1552
Query: 374 QEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPIVVTASSDLSLLGFGINAENN 433
+ +QQ+E+ +V++YRA LS D++VIPAG+P+VV A+SDL+ FGINAENN
Sbjct: 1553 R--------QQQEEQPLEVRKYRAELSEQDIFVIPAGYPVVVNATSDLNFFAFGINAENN 1708
Query: 434 QRNFLA 439
QRNFLA
Sbjct: 1709 QRNFLA 1726
>TC225160 UP|Q948X9 (Q948X9) Beta-conglycinin alpha-subunit, complete
Length = 2152
Score = 400 bits (1028), Expect = e-112
Identities = 198/366 (54%), Positives = 283/366 (77%)
Frame = +3
Query: 74 ENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHND 133
+NPF+F SNRF+TLF+N+ G IR+LQRF++RS +NL++YR+LE++SKP+TL LP H D
Sbjct: 705 KNPFLFGSNRFETLFKNQYGRIRVLQRFNQRSPQLQNLRDYRILEFNSKPNTLLLPNHAD 884
Query: 134 ADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDLRVLDLAIP 193
AD+++V+++G AIL+++N ++R+S+ L+ GD +++P+GT Y+ N D+N++LR++ LAIP
Sbjct: 885 ADYLIVILNGTAILSLVNNDDRDSYRLQSGDALRVPSGTTYYVVNPDNNENLRLITLAIP 1064
Query: 194 VNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEPQHRRGLR 253
VN+PG+F+SF LS +E QQS+L GFS+NILEA++++ +EEI +VL E G +
Sbjct: 1065 VNKPGRFESFFLSSTEAQQSYLQGFSRNILEASYDTKFEEINKVLFSREE-------GQQ 1223
Query: 254 KDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQKPIYSNKFGNF 313
+ E+R Q +VIV++S+EQI LSK AKSSSR++ SSE +P NLR++ PIYSNK G F
Sbjct: 1224 QGEQRLQE---SVIVEISKEQIRALSKRAKSSSRKTISSEDKPFNLRSRDPIYSNKLGKF 1394
Query: 314 FEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQ 373
FEITPEKNPQL+DLDI ++ ++ EG+LLLPHFNS+A VI+ + EG ELVG + E Q
Sbjct: 1395 FEITPEKNPQLRDLDIFLSIVDMNEGALLLPHFNSKAIVILVINEGDANIELVGLK-EQQ 1571
Query: 374 QEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPIVVTASSDLSLLGFGINAENN 433
QEQ QQ+E+ +V++YRA LS D++VIPAG+P+VV A+S+L+ GINAENN
Sbjct: 1572 QEQ-------QQEEQPLEVRKYRAELSEQDIFVIPAGYPVVVNATSNLNFFAIGINAENN 1730
Query: 434 QRNFLA 439
QRNFLA
Sbjct: 1731 QRNFLA 1748
Score = 32.3 bits (72), Expect = 0.44
Identities = 40/186 (21%), Positives = 80/186 (42%), Gaps = 15/186 (8%)
Frame = +3
Query: 240 EENEQEPQHRRGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSES--EPI 297
+E+E++ + + + +++S+E R+Q E+ + + S SESSES E
Sbjct: 537 DEDEEQDERQFPFPRPPHQKESEE--------RKQEEDEDEEQQRESEESESSESQRELR 692
Query: 298 NLRNQKP----------IYSNKFGNF--FEITPEKNPQLKDL-DILVNYAEIREGSLLLP 344
+N+ P ++ N++G + +++PQL++L D + + +LLLP
Sbjct: 693 RHKNKNPFLFGSNRFETLFKNQYGRIRVLQRFNQRSPQLQNLRDYRILEFNSKPNTLLLP 872
Query: 345 HFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQQQERSQQVQRYRARLSPGDV 404
+ ++ A ++ + G LV N ++ Y RL GD
Sbjct: 873 N-HADADYLIVILNGTAILSLV-----NNDDRDSY------------------RLQSGDA 980
Query: 405 YVIPAG 410
+P+G
Sbjct: 981 LRVPSG 998
>TC211438 UP|GLCX_SOYBN (P11827) Beta-conglycinin, alpha' chain precursor,
complete
Length = 1959
Score = 382 bits (981), Expect = e-106
Identities = 192/384 (50%), Positives = 283/384 (73%), Gaps = 19/384 (4%)
Frame = +1
Query: 75 NPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHNDA 134
NPF FNS RFQTLF+N+ GH+R+LQRF+KRS+ +NL++YR+LE++SKP+TL LP H DA
Sbjct: 667 NPFHFNSKRFQTLFKNQYGHVRVLQRFNKRSQQLQNLRDYRILEFNSKPNTLLLPHHADA 846
Query: 135 DFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLP-------------------AGTLGY 175
D+++V+++G AILT++N ++R+S+NL+ GD +++P AGT Y
Sbjct: 847 DYLIVILNGTAILTLVNNDDRDSYNLQSGDALRVPAGTTFYVVNPDNDENLRMIAGTTFY 1026
Query: 176 LANRDDNKDLRVLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIE 235
+ N D++++LR++ LAIPVN+PG+F+SF LS ++ QQS+L GFSKNILEA++++ +EEI
Sbjct: 1027 VVNPDNDENLRMITLAIPVNKPGRFESFFLSSTQAQQSYLQGFSKNILEASYDTKFEEIN 1206
Query: 236 RVLIEENEQEPQHRRGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESE 295
+VL E + Q +ER Q+S VIV++S++QI ELSK+AKSSSR++ SSE +
Sbjct: 1207 KVLFGREEGQQQ------GEERLQES----VIVEISKKQIRELSKHAKSSSRKTISSEDK 1356
Query: 296 PINLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVA 355
P NL ++ PIYSNK G FEIT ++NPQL+DLD+ ++ ++ EG+L LPHFNS+A V++
Sbjct: 1357 PFNLGSRDPIYSNKLGKLFEIT-QRNPQLRDLDVFLSVVDMNEGALFLPHFNSKAIVVLV 1533
Query: 356 VEEGKGEFELVGQRNENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPIVV 415
+ EG+ ELVG + + Q+ +QQ+E+ +V++YRA LS D++VIPAG+P++V
Sbjct: 1534 INEGEANIELVGIKEQQQR--------QQQEEQPLEVRKYRAELSEQDIFVIPAGYPVMV 1689
Query: 416 TASSDLSLLGFGINAENNQRNFLA 439
A+SDL+ FGINAENNQRNFLA
Sbjct: 1690 NATSDLNFFAFGINAENNQRNFLA 1761
>TC203543 similar to UP|O22121 (O22121) Beta subunit of beta conglycinin
(Fragment), partial (92%)
Length = 1421
Score = 381 bits (979), Expect = e-106
Identities = 197/367 (53%), Positives = 267/367 (72%)
Frame = +3
Query: 73 QENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHN 132
Q NPF F+SNRFQTLF+N++GH+R+LQRFD+RS ENL++YR++E +KP+TLFLP H
Sbjct: 87 QNNPFHFSSNRFQTLFKNQHGHLRVLQRFDQRSPQLENLRDYRVVELMAKPNTLFLPHHA 266
Query: 133 DADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDLRVLDLAI 192
DADF+L+V+SG+A++ ++ P++R+ + L+RG ++PAGT YL N D KDLRV+ LAI
Sbjct: 267 DADFLLLVLSGRALINLVKPDDRDPYYLDRGYAQRIPAGTTVYLVNPDKKKDLRVIKLAI 446
Query: 193 PVNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEPQHRRGL 252
PVN+PG F+ F LS +++QQS+L GFS+NILEA+FN+ +EEI RVL
Sbjct: 447 PVNKPGNFEDFFLSSTQDQQSYLQGFSENILEASFNTKFEEINRVLF------------- 587
Query: 253 RKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQKPIYSNKFGN 312
E R+ QE VI+++S+EQI ELSK AKSSSR + S + EP LR + I SN FG
Sbjct: 588 -GGEGRRHQQE-GVILELSKEQIRELSKRAKSSSRSTNSFDYEPFYLRGSQ-ISSNNFGK 758
Query: 313 FFEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNEN 372
F+EITPEKNPQL+D DIL+N +I EG LLLPH+NS+A VI+ V EG+ ELVG +
Sbjct: 759 FYEITPEKNPQLRDFDILLNTVDINEGGLLLPHYNSKAIVILMVTEGEANIELVGLK--- 929
Query: 373 QQEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPIVVTASSDLSLLGFGINAEN 432
++QQ E +++V++YRA LS D++VIPA +P VV A+S+L+ + FGINAEN
Sbjct: 930 ---------EQQQGEETREVRKYRAELSEDDIFVIPAAYPFVVNATSNLNFVAFGINAEN 1082
Query: 433 NQRNFLA 439
NQRNFLA
Sbjct: 1083NQRNFLA 1103
>TC203614 UP|Q94LX2 (Q94LX2) Beta-conglycinin alpha subunit, complete
Length = 1983
Score = 380 bits (977), Expect = e-106
Identities = 196/370 (52%), Positives = 276/370 (73%), Gaps = 4/370 (1%)
Frame = +3
Query: 74 ENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYH----SKPHTLFLP 129
+NPF+F SNRF+TLF+N+ G IR+LQRF++R F LQN L + SKP+TL LP
Sbjct: 615 KNPFLFGSNRFETLFKNQYGRIRVLQRFNQR---FPQLQNLPRLPHFWSSTSKPNTLLLP 785
Query: 130 QHNDADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDLRVLD 189
H DAD+++V+++G AIL+++N ++R+S+ L+ GD +++P+GT Y+ N D+N++LR++
Sbjct: 786 NHADADYLIVILNGTAILSLVNNDDRDSYRLQSGDALRVPSGTTYYVVNPDNNENLRLIT 965
Query: 190 LAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEPQHR 249
LAIPVN+PG+F+SF LS +E QQS+L GFS+NILEA++++ +EEI +VL E + Q
Sbjct: 966 LAIPVNKPGRFESFFLSSTEAQQSYLQGFSRNILEASYDTKFEEINKVLFSREEGQQQG- 1142
Query: 250 RGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQKPIYSNK 309
++R Q+S VIV++S+EQI LSK AKSSSR++ SSE +P NLR++ PIYSNK
Sbjct: 1143-----EQRLQES----VIVEISKEQIRALSKRAKSSSRKTISSEDKPFNLRSRDPIYSNK 1295
Query: 310 FGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQR 369
G FFEITPEKNPQL+DLDI ++ ++ EG+LLLPHFNS+A VI+ + EG ELVG +
Sbjct: 1296LGKFFEITPEKNPQLRDLDIFLSIVDMNEGALLLPHFNSKAIVILVINEGDANIELVGLK 1475
Query: 370 NENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPIVVTASSDLSLLGFGIN 429
E QQEQ QQ+E+ +V++YRA LS D++VIPAG+P+VV A+S+L+ GIN
Sbjct: 1476-EQQQEQ-------QQEEQPLEVRKYRAELSEQDIFVIPAGYPVVVNATSNLNFFAIGIN 1631
Query: 430 AENNQRNFLA 439
AENNQRNFLA
Sbjct: 1632AENNQRNFLA 1661
Score = 29.6 bits (65), Expect = 2.8
Identities = 35/174 (20%), Positives = 69/174 (39%), Gaps = 17/174 (9%)
Frame = +3
Query: 254 KDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQK---------- 303
+DE +Q + + ++ E + + ++ ES ESE LR K
Sbjct: 462 EDEDEEQDERQFPFPRPPHQKEERKQEEDEDEEQQRESEESEDSELRRHKNKNPFLFGSN 641
Query: 304 ---PIYSNKFGNF--FEITPEKNPQLKDLDILVNY--AEIREGSLLLPHFNSRATVIVAV 356
++ N++G + ++ PQL++L L ++ + + +LLLP+ ++ A ++ +
Sbjct: 642 RFETLFKNQYGRIRVLQRFNQRFPQLQNLPRLPHFWSSTSKPNTLLLPN-HADADYLIVI 818
Query: 357 EEGKGEFELVGQRNENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAG 410
G LV N ++ Y RL GD +P+G
Sbjct: 819 LNGTAILSLV-----NNDDRDSY------------------RLQSGDALRVPSG 911
>TC203890 homologue to UP|Q7XXT2 (Q7XXT2) Prepro beta-conglycinin alpha prime
subunit, partial (36%)
Length = 742
Score = 228 bits (582), Expect = 3e-60
Identities = 114/253 (45%), Positives = 181/253 (71%)
Frame = +2
Query: 74 ENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHND 133
+NPF FNS R QTL E++ G F R NL++YR+LE++SKP+TL P H D
Sbjct: 14 KNPFHFNSKRLQTLLESKYGLP*ASWDFIHRVLELPNLRDYRILEFNSKPNTLLFPHHAD 193
Query: 134 ADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDLRVLDLAIP 193
AD+++++++G AILT++N +NR+S+NL+ GD +++PAGT Y+ N D++++LR++ LAIP
Sbjct: 194 ADYLIIILNGTAILTLVNNDNRDSYNLQSGDALRVPAGTTYYVVNPDNDENLRMITLAIP 373
Query: 194 VNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEPQHRRGLR 253
VN+PG+F+SF LS ++ QQS+L GFSKNILEA++++ +EEI + L E + Q
Sbjct: 374 VNKPGRFESFFLSSTQAQQSYLQGFSKNILEASYDTKFEEINKGLFGREEGQQQ------ 535
Query: 254 KDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQKPIYSNKFGNF 313
+ER Q+S VIV++S++Q+ + K++ + +R++ SE +P +LR++ PIY NK G
Sbjct: 536 GEERLQES----VIVEISKKQMRDRGKHSGTRARKTIFSEDKPCDLRSRDPIYCNKLGKL 703
Query: 314 FEITPEKNPQLKD 326
EITPEK+P+L++
Sbjct: 704 LEITPEKSPELQN 742
>TC225159 homologue to UP|O22120 (O22120) Alpha subunit of beta conglycinin
(Fragment), partial (50%)
Length = 849
Score = 209 bits (533), Expect = 2e-54
Identities = 103/229 (44%), Positives = 165/229 (71%)
Frame = +1
Query: 74 ENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHND 133
+NPF+F SNRF+TLF+N+ G IR+LQRF++RS +NL++YR+LE++SKP+TL LP H D
Sbjct: 166 KNPFLFGSNRFETLFKNQYGRIRVLQRFNQRSPQLQNLRDYRILEFNSKPNTLLLPNHAD 345
Query: 134 ADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDLRVLDLAIP 193
AD+++V+++G AIL+++N ++R+S+ L+ GD +++P+GT Y+ N D+N++LR++ LAIP
Sbjct: 346 ADYLIVILNGTAILSLVNNDDRDSYRLQSGDALRVPSGTTYYVVNPDNNENLRLITLAIP 525
Query: 194 VNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEPQHRRGLR 253
N+PG+F+SF LS +E QQS+L GFS NILEA++++ ++EI + L+ E + Q
Sbjct: 526 DNKPGRFESFFLSSTEAQQSYLQGFSMNILEASYDTKFKEINKDLLSREEGQQQ------ 687
Query: 254 KDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQ 302
QQ + +VI +S+EQ LS+ +S R+ SE LR++
Sbjct: 688 ----GQQRLQESVIDDISKEQNTALSRRRQSXLMRTILSEDIHFYLRSR 822
Score = 30.4 bits (67), Expect = 1.7
Identities = 35/173 (20%), Positives = 68/173 (39%), Gaps = 16/173 (9%)
Frame = +1
Query: 254 KDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQK---------- 303
+DE +Q + + ++ E + + ++ ES ESE LR K
Sbjct: 13 EDEDEEQDERQFPFPRPPHQKEERKQEEDEDEEQQRESEESEDSELRRHKNKNPFLFGSN 192
Query: 304 ---PIYSNKFGNF--FEITPEKNPQLKDL-DILVNYAEIREGSLLLPHFNSRATVIVAVE 357
++ N++G + +++PQL++L D + + +LLLP+ ++ A ++ +
Sbjct: 193 RFETLFKNQYGRIRVLQRFNQRSPQLQNLRDYRILEFNSKPNTLLLPN-HADADYLIVIL 369
Query: 358 EGKGEFELVGQRNENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAG 410
G LV N ++ Y RL GD +P+G
Sbjct: 370 NGTAILSLV-----NNDDRDSY------------------RLQSGDALRVPSG 459
>TC217163 UP|Q84V19 (Q84V19) Sucrose-binding protein 2, complete
Length = 1722
Score = 199 bits (506), Expect = 2e-51
Identities = 112/370 (30%), Positives = 199/370 (53%), Gaps = 4/370 (1%)
Frame = +1
Query: 70 DQDQENPFIFNSNR-FQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFL 128
+++ ENP++F ++ F T E E G IR+L++F ++SK+ + ++N+RL ++ HT
Sbjct: 301 EEEDENPYVFEEDKDFSTRVETEGGSIRVLKKFTEKSKLLQGIENFRLAILEARAHTFVS 480
Query: 129 PQHNDADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDLRVL 188
P+H D++ +L + G+A+L ++ + LE GD I +PAGT Y+ NRD+N+ L +
Sbjct: 481 PRHFDSEVVLFNIKGRAVLGLVRESETEKITLEPGDMIHIPAGTPLYIVNRDENEKLLLA 660
Query: 189 DLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEPQH 248
L IPV+ PG+F+ F + +S LS FS N+L+AA + ++ER+ ++NE
Sbjct: 661 MLHIPVSTPGKFEEFFGPGGRDPESVLSAFSWNVLQAALQTPKGKLERLFNQQNE----- 825
Query: 249 RRGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQKPIYSN 308
I K+SRE++ L+ KSS N+ +++P +SN
Sbjct: 826 ----------------GSIFKISRERVRALAPTKKSSWWPFGGESKAQFNIFSKRPTFSN 957
Query: 309 KFGNFFEITP-EKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVG 367
+G E+ P ++ L+ L++++ + I + S+ H+NS AT I V +G+G ++
Sbjct: 958 GYGRLTEVGPDDEKSWLQRLNLMLTFTNITQRSMSTIHYNSHATKIALVMDGRGHLQISC 1137
Query: 368 QRNENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPIVVTASS--DLSLLG 425
++ D + + S R A L PG V+V+P GHP V AS+ +L ++
Sbjct: 1138PHMSSR-------SDSKHDKSSPSYHRISADLKPGMVFVVPPGHPFVTIASNKENLLIIC 1296
Query: 426 FGINAENNQR 435
F +N +N++
Sbjct: 1297FEVNVRDNKK 1326
>TC203891 homologue to UP|GLCA_SOYBN (P13916) Beta-conglycinin, alpha chain
precursor, partial (36%)
Length = 663
Score = 198 bits (504), Expect = 4e-51
Identities = 104/178 (58%), Positives = 134/178 (74%)
Frame = +1
Query: 262 QEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQKPIYSNKFGNFFEITPEKN 321
++ NVIV++S+EQI LSK AKSSSR++ SSE +P NLR++ PIYSNK G FFEITPEKN
Sbjct: 94 RKKNVIVEISKEQIRALSKRAKSSSRKTISSEDKPFNLRSRDPIYSNKLGKFFEITPEKN 273
Query: 322 PQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEE 381
PQL+DLDI ++ ++ EG+LLLPHFNS+A VI+ + EG ELVG + E QQEQ
Sbjct: 274 PQLRDLDIFLSIVDMNEGALLLPHFNSKAIVILVINEGDANIELVGLK-EQQQEQ----- 435
Query: 382 DEQQQERSQQVQRYRARLSPGDVYVIPAGHPIVVTASSDLSLLGFGINAENNQRNFLA 439
QQ+E+ +V++YR DV+VIPA +P VV A+S+L+ L FGINAENNQRNFLA
Sbjct: 436 --QQEEQPLEVRKYR-----DDVFVIPAAYPFVVNATSNLNFLAFGINAENNQRNFLA 588
>TC204732 homologue to UP|SBP_SOYBN (Q04672) Sucrose-binding protein
precursor (SBP), complete
Length = 1850
Score = 189 bits (480), Expect = 2e-48
Identities = 110/373 (29%), Positives = 199/373 (52%), Gaps = 7/373 (1%)
Frame = +2
Query: 70 DQDQENPFIFNSNR-FQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFL 128
++ ENP+IF ++ F+T E E G IR+L++F ++SK+ + ++N+RL ++ HT
Sbjct: 362 EEQDENPYIFEEDKDFETRVETEGGRIRVLKKFTEKSKLLQGIENFRLAILEARAHTFVS 541
Query: 129 PQHNDADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDLRVL 188
P+H D++ + + G+A+L +++ + LE GD I +PAGT Y+ NRD+N L +
Sbjct: 542 PRHFDSEVVFFNIKGRAVLGLVSESETEKITLEPGDMIHIPAGTPLYIVNRDENDKLFLA 721
Query: 189 DLAIP--VNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEP 246
L IP V+ PG+F+ F + +S LS FS N+L+AA + ++E++ ++NE
Sbjct: 722 MLHIPVSVSTPGKFEEFFGPGGRDPESVLSAFSWNVLQAALQTPKGKLEKLFDQQNE--- 892
Query: 247 QHRRGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQKPIY 306
I +SREQ+ L+ KSS N+ +++P
Sbjct: 893 ------------------GSIFAISREQVRALAPTKKSSWWPFGGESKPQFNIFSKRPTI 1018
Query: 307 SNKFGNFFEITPEKNPQ--LKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFE 364
SN +G E+ P+ + + L+ L++++ + I + S+ H+NS AT I V +G+G +
Sbjct: 1019SNGYGRLTEVGPDDDEKSWLQRLNLMLTFTNITQRSMSTIHYNSHATKIALVIDGRGHLQ 1198
Query: 365 LVGQRNENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPIVVTASS--DLS 422
+ ++ ++++ S R + L PG V+V+P GHP V AS+ +L
Sbjct: 1199ISCPHMSSRSSHSKHDKS------SPSYHRISSDLKPGMVFVVPPGHPFVTIASNKENLL 1360
Query: 423 LLGFGINAENNQR 435
++ F +NA +N++
Sbjct: 1361MICFEVNARDNKK 1399
>AW568367 similar to GP|15425635|db beta-conglycinin beta-subunit {Glycine
max}, partial (43%)
Length = 568
Score = 180 bits (456), Expect = 1e-45
Identities = 89/183 (48%), Positives = 132/183 (71%), Gaps = 1/183 (0%)
Frame = +2
Query: 53 MLLGIVFLASICVSSRSDQDQENPFIFN-SNRFQTLFENENGHIRLLQRFDKRSKIFENL 111
+LLG FLAS CV + D+ NPF S+ FQTLF+N+NG I LLQRF +R ENL
Sbjct: 17 VLLGTGFLASFCVYLQVR*DENNPFYLRTSSTFQTLFDNQNGRIPLLQRFTERFPHLENL 196
Query: 112 QNYRLLEYHSKPHTLFLPQHNDADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAG 171
+YR+++ HS+P+T+ LP H DADF+L V++G+AILT++N ++R+ +NL GD ++PAG
Sbjct: 197 TDYRIVQCHSQPNTILLPHHADADFLLFVLTGRAILTLVNNDDRDFYNLHPGDAQRIPAG 376
Query: 172 TLGYLANRDDNKDLRVLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNY 231
T YL N D+++L+++ LAI V++PG++ F LS ++ QQS+L FS NILE + +S +
Sbjct: 377 TTYYLVNPHDHQNLKIIKLAIHVDKPGRYDDFFLSSTQAQQSYLQSFSHNILETSCHSEF 556
Query: 232 EEI 234
EE+
Sbjct: 557 EEM 565
>AW317839 similar to PIR|S20007|S20 beta-conglycinin alpha chain precursor -
soybean, partial (29%)
Length = 541
Score = 159 bits (403), Expect = 2e-39
Identities = 87/190 (45%), Positives = 122/190 (63%)
Frame = +2
Query: 158 FNLERGDTIKLPAGTLGYLANRDDNKDLRVLDLAIPVNRPGQFQSFSLSESENQQSFLSG 217
+ L+ GD +++P+GT Y+ N DDN+ LR++ LAIP N+PG F+SF LS +E QQS+L G
Sbjct: 2 YRLQSGDALRVPSGTTYYVVNPDDNEYLRLITLAIPANKPGGFESFFLSSTEAQQSYLQG 181
Query: 218 FSKNILEAAFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQEANVIVKVSREQIEE 277
++ NIL A +++ +EEI VL E + +R +Q +VIV++S+EQI
Sbjct: 182 YTMNIL*ACYDTKFEEINEVLCT*EEGQ----------QRGEQRLP*SVIVQISKEQIRA 331
Query: 278 LSKNAKSSSRRSESSESEPINLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIR 337
LSK A SSSRR+ S EP NLR + IYSNK G ITPEK Q +D DI ++ ++
Sbjct: 332 LSKRA*SSSRRTMGS*DEPFNLRRRDLIYSNKLGKCCWITPEKYRQHRDYDIYLSIVDMY 511
Query: 338 EGSLLLPHFN 347
EG+ LPHFN
Sbjct: 512 EGATTLPHFN 541
>AI941229 similar to PIR|S20007|S20 beta-conglycinin alpha chain precursor -
soybean, partial (30%)
Length = 559
Score = 151 bits (382), Expect = 5e-37
Identities = 75/148 (50%), Positives = 103/148 (68%)
Frame = +2
Query: 292 SESEPINLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRAT 351
SE +P NLR++ PIYSNK FFEITPEKNPQL+D DI + ++ EG+++LPHFNS+A
Sbjct: 2 SEDKPFNLRSRYPIYSNKLAKFFEITPEKNPQLRDSDIFLIIVDMNEGAVVLPHFNSKAI 181
Query: 352 VIVAVEEGKGEFELVGQRNENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGH 411
VI+ + EG EL +E ++++QQ E+ +V+RYRA LS D++V PAG+
Sbjct: 182 VILVINEGDANIEL--------DVLKEQQQEQQQDEQPLEVRRYRAELSEQDIFVEPAGY 337
Query: 412 PIVVTASSDLSLLGFGINAENNQRNFLA 439
P VV A+S L++ INA+NNQRNFLA
Sbjct: 338 PGVVNATSYLNIFAIAINADNNQRNFLA 421
>AI941164 similar to PIR|S20007|S20 beta-conglycinin alpha chain precursor -
soybean, partial (21%)
Length = 391
Score = 120 bits (300), Expect = 2e-27
Identities = 59/127 (46%), Positives = 91/127 (71%)
Frame = +2
Query: 110 NLQNYRLLEYHSKPHTLFLPQHNDADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLP 169
N Q YRL E ++KP+TL L H DAD+++V+++G AIL+++N N+R+ + L GD ++ P
Sbjct: 11 NPQYYRLSENNAKPNTLLLHNHADADYLIVILNGTAILSLVNNNHRDCYTL*CGDALRDP 190
Query: 170 AGTLGYLANRDDNKDLRVLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNS 229
+ T Y+ N D +LRV+ LAIP N+PG SF LS +E QQS+L GFS+NILEA++++
Sbjct: 191 SRTTYYVVNPDIT*NLRVVALAIPRNKPGTVDSFFLSSTEAQQSYLQGFSRNILEASYDT 370
Query: 230 NYEEIER 236
++E +
Sbjct: 371 TFQETNK 391
>AW397351 similar to PIR|S20007|S20 beta-conglycinin alpha chain precursor -
soybean, partial (25%)
Length = 472
Score = 117 bits (292), Expect = 1e-26
Identities = 63/125 (50%), Positives = 84/125 (66%)
Frame = +1
Query: 315 EITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQ 374
EITPEKNPQL+DLDI + ++ +G+LLLPHFNS+A VI+A+ EG EL G + E QQ
Sbjct: 1 EITPEKNPQLRDLDIFLRIGDMNKGALLLPHFNSKAIVILAINEGDANIELGGLK-EQQQ 177
Query: 375 EQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPIVVTASSDLSLLGFGINAENNQ 434
EQ QQ+E+ + ++YRA S D+ V PA P V A+++L + G+NAENNQ
Sbjct: 178 EQ-------QQEEQPAEERKYRAETSEQDICVSPARDPGVDNATANLKCVAMGMNAENNQ 336
Query: 435 RNFLA 439
RNF A
Sbjct: 337 RNFRA 351
>AW509505
Length = 397
Score = 114 bits (284), Expect = 1e-25
Identities = 52/84 (61%), Positives = 71/84 (83%)
Frame = +2
Query: 81 SNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHNDADFILVV 140
SN FQTLFEN+NG IRLLQRF+KRS ENL++YR++++ SKP+T+ LP H DADF+L V
Sbjct: 146 SNSFQTLFENQNGRIRLLQRFNKRSPQLENLRDYRIVQFQSKPNTILLPHHADADFLLFV 325
Query: 141 VSGKAILTVLNPNNRNSFNLERGD 164
+SG+AILT++N ++R+S+NL GD
Sbjct: 326 LSGRAILTLVNNDDRDSYNLHPGD 397
>TC203531 UP|GLCA_SOYBN (P13916) Beta-conglycinin, alpha chain precursor,
partial (26%)
Length = 706
Score = 110 bits (275), Expect = 1e-24
Identities = 57/101 (56%), Positives = 74/101 (72%)
Frame = +1
Query: 339 GSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQQQERSQQVQRYRAR 398
G+LLLPHFNS+A VI+ + EG ELVG + E QQEQ QQ+E+ +V++YRA
Sbjct: 7 GALLLPHFNSKAIVILVINEGDANIELVGLK-EQQQEQ-------QQEEQPLEVRKYRAE 162
Query: 399 LSPGDVYVIPAGHPIVVTASSDLSLLGFGINAENNQRNFLA 439
LS D++VIPAG+P+VV A+S+L+ GINAENNQRNFLA
Sbjct: 163 LSEQDIFVIPAGYPVVVNATSNLNFFAIGINAENNQRNFLA 285
>CK769080
Length = 453
Score = 109 bits (272), Expect = 3e-24
Identities = 61/127 (48%), Positives = 87/127 (68%), Gaps = 4/127 (3%)
Frame = +3
Query: 207 ESENQQSFLSGFSK----NILEAAFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQ 262
E+ F++G+ + ILEA++++ +EEI +VL E + Q +ER Q+S
Sbjct: 99 ETLKSTGFVNGYGECYHSEILEASYDTKFEEINKVLFGREEGQQQG------EERLQES- 257
Query: 263 EANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQKPIYSNKFGNFFEITPEKNP 322
VIV++S++QI ELSK+AKSSSR++ SSE +P NLR++ PIYSNK G FEITP KNP
Sbjct: 258 ---VIVEISKKQIRELSKHAKSSSRKTISSEDKPFNLRSRVPIYSNKLGKLFEITP*KNP 428
Query: 323 QLKDLDI 329
QL+DLD+
Sbjct: 429 QLRDLDV 449
Score = 62.0 bits (149), Expect = 5e-10
Identities = 28/44 (63%), Positives = 39/44 (88%)
Frame = -2
Query: 183 KDLRVLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILEAA 226
++LR++ LAIPVN+PG+F+SF LS ++ QQS+L GFSKNILEA+
Sbjct: 161 ENLRMITLAIPVNKPGRFESFFLSSTQAQQSYLQGFSKNILEAS 30
>AW459911 similar to GP|15425631|db beta-conglycinin alpha prime subunit
{Glycine max}, partial (16%)
Length = 304
Score = 108 bits (270), Expect = 5e-24
Identities = 49/101 (48%), Positives = 78/101 (76%)
Frame = +2
Query: 86 TLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHNDADFILVVVSGKA 145
TLF N+ GH+R+LQRF+KRS+ +NL++YR+LE + KP+T+ +P H D+D+++V+++G A
Sbjct: 2 TLFRNQYGHVRVLQRFNKRSQQLQNLRDYRILELNFKPNTVLVPHHADSDYLIVILNGTA 181
Query: 146 ILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDLR 186
ILT++N ++R+S+ LE GD + PAGT Y+ N D + LR
Sbjct: 182 ILTLVNNDDRDSYKLESGDAL*GPAGTTYYVVNPDYD*SLR 304
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.133 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,727,620
Number of Sequences: 63676
Number of extensions: 191159
Number of successful extensions: 1391
Number of sequences better than 10.0: 108
Number of HSP's better than 10.0 without gapping: 1325
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1349
length of query: 440
length of database: 12,639,632
effective HSP length: 100
effective length of query: 340
effective length of database: 6,272,032
effective search space: 2132490880
effective search space used: 2132490880
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC148289.2


