

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148217.3 - phase: 2
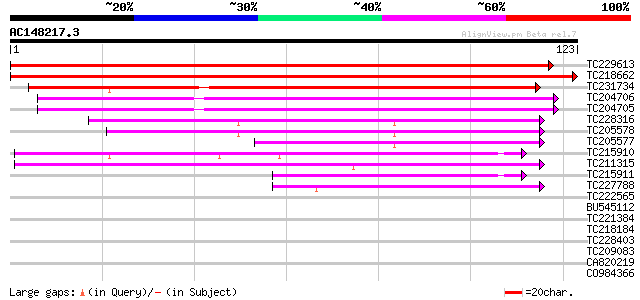
(123 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC229613 weakly similar to UP|Q9MBD6 (Q9MBD6) Ntdin, partial (40%) 207 1e-54
TC218662 similar to UP|Q9FKW8 (Q9FKW8) Senescence-associated pro... 181 7e-47
TC231734 weakly similar to UP|Q7Y234 (Q7Y234) At2g21045, partial... 121 9e-29
TC204706 similar to UP|Q9MBD6 (Q9MBD6) Ntdin, partial (61%) 100 2e-22
TC204705 similar to UP|Q9MBD6 (Q9MBD6) Ntdin, partial (61%) 99 4e-22
TC228316 similar to UP|Q94A65 (Q94A65) AT4g27700/T29A15_190, par... 50 3e-07
TC205578 similar to UP|Q94A65 (Q94A65) AT4g27700/T29A15_190, par... 45 6e-06
TC205577 similar to UP|Q94A65 (Q94A65) AT4g27700/T29A15_190, par... 42 5e-05
TC215910 similar to UP|O48529 (O48529) Rhodanese-like family pro... 42 9e-05
TC211315 similar to UP|Q9LMU3 (Q9LMU3) F2H15.8 protein, partial ... 41 1e-04
TC215911 similar to UP|Q6H444 (Q6H444) Rhodanese family protein-... 39 5e-04
TC227788 weakly similar to UP|Q8LBR6 (Q8LBR6) Rhodanese-like fam... 39 5e-04
TC222565 similar to UP|Q7XJN9 (Q7XJN9) At2g40760 protein, partia... 36 0.005
BU545112 33 0.043
TC221384 similar to UP|Q9LMU3 (Q9LMU3) F2H15.8 protein, partial ... 33 0.043
TC218184 similar to UP|O23727 (O23727) Peptidyl-prolyl cis-trans... 33 0.043
TC228403 similar to UP|Q6ZI49 (Q6ZI49) Rhodanese-like domain-con... 32 0.073
TC209083 homologue to GB|AAL15278.1|16323087|AY057647 AT3g01490/... 28 1.4
CA820219 weakly similar to GP|20197120|gb| expressed protein {Ar... 27 1.8
CO984366 27 3.1
>TC229613 weakly similar to UP|Q9MBD6 (Q9MBD6) Ntdin, partial (40%)
Length = 872
Score = 207 bits (526), Expect = 1e-54
Identities = 100/118 (84%), Positives = 107/118 (89%)
Frame = +2
Query: 1 SKTEVVTVDVLATKSLIKTTHVYLDVRTVEEFQKGHVDSEKIINIAYMFNTPEGRVKNPE 60
S EVVTVDV ATK LI+T+HVYLDVRTVEEFQKGHVD+EKIIN+AYMFNTPEGRVKNPE
Sbjct: 212 SGPEVVTVDVHATKDLIQTSHVYLDVRTVEEFQKGHVDAEKIINVAYMFNTPEGRVKNPE 391
Query: 61 FLKEVSSLCNKEDHLIVGCQSGVRSVYATADLLAEGFKDVYNMGGGYLEWVKKEFPVK 118
FLKEVS C KEDH+IVGCQSGVRS+YATADLL EGFKDV NMGGGYL+WVK E PVK
Sbjct: 392 FLKEVSYACKKEDHIIVGCQSGVRSLYATADLLTEGFKDVSNMGGGYLDWVKNELPVK 565
>TC218662 similar to UP|Q9FKW8 (Q9FKW8) Senescence-associated protein
sen1-like protein (AT5g66170/K2A18_25), partial (18%)
Length = 941
Score = 181 bits (459), Expect = 7e-47
Identities = 85/123 (69%), Positives = 105/123 (85%)
Frame = +3
Query: 1 SKTEVVTVDVLATKSLIKTTHVYLDVRTVEEFQKGHVDSEKIINIAYMFNTPEGRVKNPE 60
S +VV +DV A K LI+T +YLDVRTVEEF+KGHVD+ ++NI YM NTP+G+VKNP+
Sbjct: 306 SGAKVVAIDVHAAKRLIQTGSIYLDVRTVEEFKKGHVDAVNVLNIPYMLNTPKGKVKNPD 485
Query: 61 FLKEVSSLCNKEDHLIVGCQSGVRSVYATADLLAEGFKDVYNMGGGYLEWVKKEFPVKIH 120
FLKEVSS CNKEDHLI+GCQSGVRS+YATADLL+EGFK+V +MGGGY++WVK +FPV I
Sbjct: 486 FLKEVSSACNKEDHLILGCQSGVRSLYATADLLSEGFKNVKDMGGGYVDWVKNKFPVIIP 665
Query: 121 VTE 123
V +
Sbjct: 666 VAK 674
>TC231734 weakly similar to UP|Q7Y234 (Q7Y234) At2g21045, partial (81%)
Length = 647
Score = 121 bits (303), Expect = 9e-29
Identities = 60/112 (53%), Positives = 80/112 (70%), Gaps = 1/112 (0%)
Frame = +1
Query: 5 VVTVDVLATKSLIKTT-HVYLDVRTVEEFQKGHVDSEKIINIAYMFNTPEGRVKNPEFLK 63
VVT+DV A K L+ ++ + YLDVR+VEEF K HV++ N+ Y+F T GRVKNP+F+
Sbjct: 76 VVTIDVHAAKDLLNSSGYRYLDVRSVEEFNKSHVENAH--NVPYVFITEAGRVKNPDFVD 249
Query: 64 EVSSLCNKEDHLIVGCQSGVRSVYATADLLAEGFKDVYNMGGGYLEWVKKEF 115
+V+++C EDHLIV C SG RS+ A+ DLL GFK++ NMGGGY WV F
Sbjct: 250 QVAAICKNEDHLIVACNSGGRSLRASVDLLDSGFKNIVNMGGGYSAWVDAGF 405
>TC204706 similar to UP|Q9MBD6 (Q9MBD6) Ntdin, partial (61%)
Length = 921
Score = 100 bits (248), Expect = 2e-22
Identities = 52/113 (46%), Positives = 65/113 (57%)
Frame = +3
Query: 7 TVDVLATKSLIKTTHVYLDVRTVEEFQKGHVDSEKIINIAYMFNTPEGRVKNPEFLKEVS 66
+V V L+ H YLDVRT EEF GH INI YMF G KN F++EVS
Sbjct: 315 SVPVRVAYELLLAGHRYLDVRTPEEFNAGHAPGA--INIPYMFRVGSGMTKNSNFIREVS 488
Query: 67 SLCNKEDHLIVGCQSGVRSVYATADLLAEGFKDVYNMGGGYLEWVKKEFPVKI 119
S KED +IVGC+ G RS+ A +DLLA GF + +M GGY W + P ++
Sbjct: 489 SNFRKEDEIIVGCELGKRSMMAASDLLAAGFTGLTDMAGGYAAWTQNGLPTEL 647
>TC204705 similar to UP|Q9MBD6 (Q9MBD6) Ntdin, partial (61%)
Length = 851
Score = 99.4 bits (246), Expect = 4e-22
Identities = 51/113 (45%), Positives = 65/113 (57%)
Frame = +3
Query: 7 TVDVLATKSLIKTTHVYLDVRTVEEFQKGHVDSEKIINIAYMFNTPEGRVKNPEFLKEVS 66
+V V L+ H YLDVRT EEF GH INI YMF G KN F++EVS
Sbjct: 303 SVPVRVAYELLLAGHRYLDVRTPEEFDAGHAPGA--INIPYMFRVGSGMTKNSNFIREVS 476
Query: 67 SLCNKEDHLIVGCQSGVRSVYATADLLAEGFKDVYNMGGGYLEWVKKEFPVKI 119
S K+D +IVGC+ G RS+ A +DLLA GF + +M GGY W + P ++
Sbjct: 477 SQFRKDDEIIVGCELGKRSMMAASDLLAAGFTGLTDMAGGYAAWTQNGLPTEL 635
>TC228316 similar to UP|Q94A65 (Q94A65) AT4g27700/T29A15_190, partial (75%)
Length = 1012
Score = 50.1 bits (118), Expect = 3e-07
Identities = 35/130 (26%), Positives = 56/130 (42%), Gaps = 31/130 (23%)
Frame = +2
Query: 18 KTTHVYLDVRTVEEFQKGHVDSEKIINIAYM-----------------FNTPEGRVKNPE 60
+ + V LDVR EF++ H + I + F G +NPE
Sbjct: 416 ENSFVLLDVRPEAEFKEAHPPGAINVQIYRLIKEWTAWDIARRAAFLFFGIFSGTEENPE 595
Query: 61 FLKEVSSLCNKEDHLIVGCQSG--------------VRSVYATADLLAEGFKDVYNMGGG 106
F+K V + +K+ +IV C SG RS+ A L+ G+ +V+++ GG
Sbjct: 596 FIKNVEAKIDKDAKIIVACTSGGTLRPSQNLPEGQQSRSLIAAYLLVLNGYTNVFHLEGG 775
Query: 107 YLEWVKKEFP 116
+W K+E P
Sbjct: 776 LYKWFKEELP 805
>TC205578 similar to UP|Q94A65 (Q94A65) AT4g27700/T29A15_190, partial (78%)
Length = 1454
Score = 45.4 bits (106), Expect = 6e-06
Identities = 32/126 (25%), Positives = 53/126 (41%), Gaps = 31/126 (24%)
Frame = +2
Query: 22 VYLDVRTVEEFQKGHVDSEKIINIAYM-----------------FNTPEGRVKNPEFLKE 64
V LDVR EF++ H + I + F G +NPEF++
Sbjct: 425 VILDVRPEAEFKEAHPPDAINVQIYRLIKEWTAWDIARRAAFAFFGIFAGTEENPEFIQS 604
Query: 65 VSSLCNKEDHLIVGCQSG--------------VRSVYATADLLAEGFKDVYNMGGGYLEW 110
V + +K +IV C +G RS+ A L+ +G+ +V+++ GG W
Sbjct: 605 VEAKLDKNAKIIVACSAGGTMKPSQNLPEGQQSRSLIAAYLLVLDGYTNVFHLEGGLYSW 784
Query: 111 VKKEFP 116
K++ P
Sbjct: 785 FKEDLP 802
>TC205577 similar to UP|Q94A65 (Q94A65) AT4g27700/T29A15_190, partial (50%)
Length = 692
Score = 42.4 bits (98), Expect = 5e-05
Identities = 22/77 (28%), Positives = 38/77 (48%), Gaps = 14/77 (18%)
Frame = +1
Query: 54 GRVKNPEFLKEVSSLCNKEDHLIVGCQSG--------------VRSVYATADLLAEGFKD 99
G +NPEF++ V + +K +IV C +G RS+ A L+ G+ +
Sbjct: 124 GTEENPEFIQSVEAKLDKNAKIIVACSAGGTMKPSQNLPEGQQSRSLIAAYLLVLNGYTN 303
Query: 100 VYNMGGGYLEWVKKEFP 116
V+++ GG W K++ P
Sbjct: 304 VFHLEGGLYSWFKEDLP 354
>TC215910 similar to UP|O48529 (O48529) Rhodanese-like family protein,
partial (82%)
Length = 1154
Score = 41.6 bits (96), Expect = 9e-05
Identities = 38/133 (28%), Positives = 58/133 (43%), Gaps = 22/133 (16%)
Frame = +3
Query: 2 KTEVVTVDVLATKSLIKTT-HVYLDVRTVEEFQKGHVDSEKIIN--IAYMFNTPEGRVK- 57
K EV V+ K L++ + LDVR +F + H+ S + + N P +K
Sbjct: 171 KAEVKYVNAEKAKELVEADGYTVLDVRDKTQFVRAHIKSCSHVPLFVENKDNDPGTIIKR 350
Query: 58 ------------------NPEFLKEVSSLCNKEDHLIVGCQSGVRSVYATADLLAEGFKD 99
NPEF++ V S E L+V CQ G+RS A + L GF++
Sbjct: 351 QLHNNFSGLFFGLPFTKPNPEFVQSVKSQFPPESKLLVVCQEGLRSAAAASKLEEAGFEN 530
Query: 100 VYNMGGGYLEWVK 112
+ + G L+ VK
Sbjct: 531 IACITSG-LQTVK 566
>TC211315 similar to UP|Q9LMU3 (Q9LMU3) F2H15.8 protein, partial (33%)
Length = 898
Score = 41.2 bits (95), Expect = 1e-04
Identities = 28/116 (24%), Positives = 53/116 (45%), Gaps = 1/116 (0%)
Frame = +3
Query: 2 KTEVVTVDVLATKSLIKTTHVYLDVRTVEEFQKGHVDSEKIINIAYMFNTPEGRVKNPEF 61
+ E + L K ++ LDVR E+ GH + ++ NT G +
Sbjct: 546 RLESINKTDLTAKDNSNRNYILLDVRNGYEWDIGHFRGAQRPSVDCFRNTSFGLSREEIT 725
Query: 62 LKEVSSLCNKED-HLIVGCQSGVRSVYATADLLAEGFKDVYNMGGGYLEWVKKEFP 116
+ S +KE ++++ C G+R ++ L +GF+++Y + GG ++K E P
Sbjct: 726 ASDPLSNVDKEKANILMYCTGGIRCDVYSSILRQQGFQNLYTLKGGVSHYLKNEGP 893
>TC215911 similar to UP|Q6H444 (Q6H444) Rhodanese family protein-like,
partial (48%)
Length = 734
Score = 39.3 bits (90), Expect = 5e-04
Identities = 23/55 (41%), Positives = 32/55 (57%)
Frame = +1
Query: 58 NPEFLKEVSSLCNKEDHLIVGCQSGVRSVYATADLLAEGFKDVYNMGGGYLEWVK 112
NPEF+ V S E L+V CQ G+RS A + L A GF+++ + G L+ VK
Sbjct: 16 NPEFVPSVKSQFPPESKLLVVCQEGLRSAAAASKLEAAGFENIACITSG-LQTVK 177
>TC227788 weakly similar to UP|Q8LBR6 (Q8LBR6) Rhodanese-like family protein,
partial (79%)
Length = 887
Score = 39.3 bits (90), Expect = 5e-04
Identities = 21/60 (35%), Positives = 35/60 (58%), Gaps = 1/60 (1%)
Frame = +1
Query: 58 NPEFLKEV-SSLCNKEDHLIVGCQSGVRSVYATADLLAEGFKDVYNMGGGYLEWVKKEFP 116
N EFL +V +++ KE L+V C G+RS+ A + L G+K++ + GG+ +FP
Sbjct: 439 NSEFLSQVENAIPGKEAKLLVACGEGLRSMTAASKLYNGGYKNLGWLAGGFNRSKNNDFP 618
>TC222565 similar to UP|Q7XJN9 (Q7XJN9) At2g40760 protein, partial (34%)
Length = 722
Score = 35.8 bits (81), Expect = 0.005
Identities = 17/47 (36%), Positives = 32/47 (67%), Gaps = 3/47 (6%)
Frame = +1
Query: 70 NKEDHL---IVGCQSGVRSVYATADLLAEGFKDVYNMGGGYLEWVKK 113
NK H+ + C G+R AT+ LL++GFK+V+++ GG L+++++
Sbjct: 232 NKRQHMPRVAMYCT*GIRCEKATSLLLSKGFKEVFHLEGGILKYLEE 372
>BU545112
Length = 461
Score = 32.7 bits (73), Expect = 0.043
Identities = 14/43 (32%), Positives = 25/43 (57%)
Frame = -1
Query: 64 EVSSLCNKEDHLIVGCQSGVRSVYATADLLAEGFKDVYNMGGG 106
E+++ + + V C G+RS+ L ++GF+ VYN+ GG
Sbjct: 374 EITTKFDPQKDTYVMCHHGMRSLQVAKWLQSQGFRKVYNLSGG 246
>TC221384 similar to UP|Q9LMU3 (Q9LMU3) F2H15.8 protein, partial (33%)
Length = 857
Score = 32.7 bits (73), Expect = 0.043
Identities = 13/54 (24%), Positives = 33/54 (61%)
Frame = +3
Query: 65 VSSLCNKEDHLIVGCQSGVRSVYATADLLAEGFKDVYNMGGGYLEWVKKEFPVK 118
+S++ ++ ++++ C G+R ++ L +GF+++Y + GG ++K E P +
Sbjct: 114 LSNVDKEKANILMYCTGGIRCDVYSSILRQQGFQNLYTLKGGVSHYLKNEGPAE 275
>TC218184 similar to UP|O23727 (O23727) Peptidyl-prolyl cis-trans isomerase ,
partial (92%)
Length = 1203
Score = 32.7 bits (73), Expect = 0.043
Identities = 14/43 (32%), Positives = 25/43 (57%)
Frame = +3
Query: 64 EVSSLCNKEDHLIVGCQSGVRSVYATADLLAEGFKDVYNMGGG 106
E+++ + + V C G+RS+ L ++GF+ VYN+ GG
Sbjct: 711 EITTKFDPQKDTYVMCHHGMRSLQVAKWLQSQGFRKVYNLSGG 839
>TC228403 similar to UP|Q6ZI49 (Q6ZI49) Rhodanese-like domain-containing
protein-like, partial (63%)
Length = 1101
Score = 32.0 bits (71), Expect = 0.073
Identities = 17/46 (36%), Positives = 27/46 (57%)
Frame = +1
Query: 60 EFLKEVSSLCNKEDHLIVGCQSGVRSVYATADLLAEGFKDVYNMGG 105
+FL +V K+ LIV CQ G+RS+ A L G+K+++ + G
Sbjct: 265 QFLAKVEEKFPKDAELIVVCQKGLRSLAACELLYNAGYKNLFWVQG 402
>TC209083 homologue to GB|AAL15278.1|16323087|AY057647 AT3g01490/F4P13_4
{Arabidopsis thaliana;} , partial (51%)
Length = 1076
Score = 27.7 bits (60), Expect = 1.4
Identities = 13/41 (31%), Positives = 23/41 (55%)
Frame = +3
Query: 41 KIINIAYMFNTPEGRVKNPEFLKEVSSLCNKEDHLIVGCQS 81
K + + +PE + ++ + L E S CN+E ++GCQS
Sbjct: 417 KFLRNHFSCRSPELKTRSTKMLPEFSGKCNEE---VLGCQS 530
>CA820219 weakly similar to GP|20197120|gb| expressed protein {Arabidopsis
thaliana}, partial (7%)
Length = 422
Score = 27.3 bits (59), Expect = 1.8
Identities = 17/52 (32%), Positives = 26/52 (49%), Gaps = 6/52 (11%)
Frame = -1
Query: 27 RTVEEFQKGHVDSEK--IINIAYMFNT----PEGRVKNPEFLKEVSSLCNKE 72
RT EEF GHVD+ K IN +Y+ + +P+ E S+C+ +
Sbjct: 188 RTAEEFSSGHVDARKHEKINASYVLQKEIT*TQCLTCHPQKF*ETMSICHMQ 33
>CO984366
Length = 732
Score = 26.6 bits (57), Expect = 3.1
Identities = 14/44 (31%), Positives = 22/44 (49%)
Frame = +2
Query: 23 YLDVRTVEEFQKGHVDSEKIINIAYMFNTPEGRVKNPEFLKEVS 66
++D + E K H ++++ Y F R KNP+ KEVS
Sbjct: 200 FIDSQKHSEIMKCHTNNKRNPEFIYKFLVSIRR*KNPDTQKEVS 331
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.135 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,940,027
Number of Sequences: 63676
Number of extensions: 56620
Number of successful extensions: 223
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 218
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 219
length of query: 123
length of database: 12,639,632
effective HSP length: 85
effective length of query: 38
effective length of database: 7,227,172
effective search space: 274632536
effective search space used: 274632536
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 53 (25.0 bits)
Medicago: description of AC148217.3


