

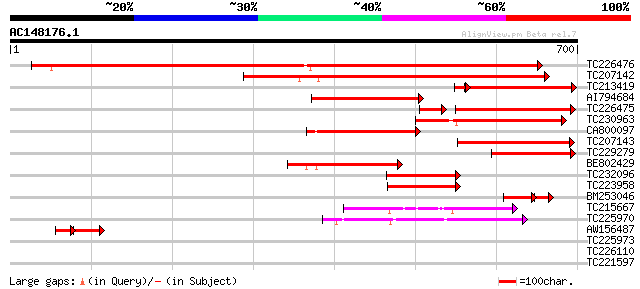
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148176.1 + phase: 0
(700 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC226476 homologue to UP|Q8L693 (Q8L693) 1-deoxy-D-xylulose 5-ph... 712 0.0
TC207142 similar to UP|Q8L692 (Q8L692) 1-deoxy-D-xylulose 5-phos... 469 e-132
TC213419 similar to UP|Q9LFL9 (Q9LFL9) 1-D-deoxyxylulose 5-phosp... 240 6e-68
AI794684 similar to GP|8953400|emb 1-D-deoxyxylulose 5-phosphate... 246 2e-65
TC226475 homologue to UP|Q8L693 (Q8L693) 1-deoxy-D-xylulose 5-ph... 211 3e-65
TC230963 similar to UP|O64904 (O64904) 1-deoxyxylulose-5-phospha... 234 1e-61
CA800097 homologue to PIR|T09543|T09 deoxyxylulose synthase (EC ... 187 2e-47
TC207143 similar to UP|Q8L692 (Q8L692) 1-deoxy-D-xylulose 5-phos... 186 3e-47
TC229279 homologue to UP|Q9XH50 (Q9XH50) 1-D-deoxyxylulose 5-pho... 153 2e-37
BE802429 similar to GP|5059160|gb| 1-D-deoxyxylulose 5-phosphate... 120 2e-27
TC232096 similar to UP|DXS_CAPAN (O78328) Probable 1-deoxy-D-xyl... 100 2e-21
TC223958 similar to UP|DXS_CAPAN (O78328) Probable 1-deoxy-D-xyl... 96 7e-20
BM253046 homologue to GP|21322713|emb 1-deoxy-D-xylulose 5-phosp... 66 2e-16
TC215667 similar to UP|ODPB_PEA (P52904) Pyruvate dehydrogenase ... 63 5e-10
TC225970 homologue to GB|AAB86804.1|2454184|ATU80186 pyruvate de... 48 2e-05
AW156487 homologue to GP|21322713|emb 1-deoxy-D-xylulose 5-phosp... 33 2e-04
TC225973 homologue to GB|AAB86804.1|2454184|ATU80186 pyruvate de... 38 0.017
TC226110 similar to UP|TKTC_CRAPL (Q42676) Transketolase, chloro... 30 4.7
TC221597 29 8.1
>TC226476 homologue to UP|Q8L693 (Q8L693) 1-deoxy-D-xylulose 5-phosphate
synthase 1 precursor, partial (85%)
Length = 2000
Score = 712 bits (1837), Expect = 0.0
Identities = 378/650 (58%), Positives = 467/650 (71%), Gaps = 18/650 (2%)
Frame = +3
Query: 27 SISQFPLSRITYSSSSRILIHRVC----SRPDIDDFYWEKVPTPILDTVQNPLCLKNLSQ 82
S SQ+ L + ++ + R C S + ++Y + PTP+LDTV P+ +KNLS
Sbjct: 57 SHSQWGLHFLAHAHRLHQMKKRPCGVYASLSESGEYYSHRPPTPLLDTVNYPIHMKNLSA 236
Query: 83 QELKQLAAEIRLELSSILSGTQILLNPSMAVVDLTVAIHHVFHAPVDKILWDVGDQTYAH 142
+ELKQLA E+R ++ +S T L S+ VV+LTVA+H+VF+AP DKILWDVG Q+Y H
Sbjct: 237 KELKQLADELRSDVIFSVSRTGGHLGSSLGVVELTVALHYVFNAPQDKILWDVGHQSYPH 416
Query: 143 KILTGRRSLMKTIRKKNGLSGFTSRFESEYDAFGAGHGCNSISAGLGMAVARDIKGRRER 202
KILTGRR M T+R+ NGLSGFT R ESE+D FG GH +ISAGLGMAV RD+KGR+
Sbjct: 417 KILTGRRDQMHTMRQTNGLSGFTKRSESEFDCFGTGHSSTTISAGLGMAVGRDLKGRKNN 596
Query: 203 VVAVISNWTTMSGQVYEAMSNAGYLDSNLVVILNDSRHSLLPKIE-DGSKTSVNALSSTL 261
VVAVI + +GQ YEAM+NAGYLDS+++VILND++ LP DG V ALSS L
Sbjct: 597 VVAVIGDGAMTAGQAYEAMNNAGYLDSDMIVILNDNKQVSLPTATLDGPIPPVGALSSAL 776
Query: 262 SRLQSSKSFRKFREAAKGVTKRIGRGMHELAAKVDEYARGMMGPPGSTLFEELGLYYIGP 321
SRLQS++ R+ RE AKGVTKRIG MHELAAKVDEYARGM+ GS+LFEELGLYYIGP
Sbjct: 777 SRLQSNRPLRELREVAKGVTKRIGGPMHELAAKVDEYARGMISGSGSSLFEELGLYYIGP 956
Query: 322 VDGHNIEDLISVLQEVASLDSMGPVLIHVITNENQVEEHNKKSYMTDK------------ 369
VDGHNI+DL+++L EV S + GPVLIHVIT + + + +K+ DK
Sbjct: 957 VDGHNIDDLVAILNEVKSTKTTGPVLIHVITEKGRGYPYAEKA--ADKYHGVTKFDPPTG 1130
Query: 370 QQDDNAGRLQTYGDCFVESLVAEAEKDKDIVVVHAGITTEPSLKLFMEKFPDRIFNVGIA 429
+Q + ++Y F E+L+AEAE DKD+V +HA + + LF +FP R F+VGIA
Sbjct: 1131KQFKSKATTRSYTTYFAEALIAEAEADKDVVAIHAAMGGGTGMNLFHRRFPTRCFDVGIA 1310
Query: 430 EQHAVTFASGLSCGGLKPFCIIPSSFLQRAYDQVVHDVDQQKVPVRFVITSAGLVGSDGP 489
EQHAVTFA+GL+C GLKPFC I SSF+QRAYDQVVHDVD QK+PVRF + AGLVG+DGP
Sbjct: 1311EQHAVTFAAGLACEGLKPFCAIYSSFMQRAYDQVVHDVDLQKLPVRFAMDRAGLVGADGP 1490
Query: 490 LQCGAFDITFMSCLPNMIVMAPSDEAELVHMVATAAHINDQPVCFRYPRGALVGKDEAIL 549
CG+FD+TFM+CLPNM+VMAPSDEAEL HMVATAA IND+P CFRYPRG +G
Sbjct: 1491THCGSFDVTFMACLPNMVVMAPSDEAELFHMVATAAAINDRPSCFRYPRGNGIGVQLPTG 1670
Query: 550 D-GIPIEIGKGRILVEGKDVALLGYGSMVQNCLKAYSLLANLGIEVTVADARFCKPLDIE 608
+ G P+EIGKGRIL+EG+ VALLGYGS QNCL A SL+ + G+ VTVADARFCKPLD
Sbjct: 1671NKGTPLEIGKGRILIEGERVALLGYGSAGQNCLPATSLVEHHGLRVTVADARFCKPLDRS 1850
Query: 609 LLRQLCKHHSFLITVEEGSIGGFGSHVAQFIALDGLLDRRIKWRPIVLPD 658
L+R L K H LIT EEGSIGGFGSHVA + + L D + P++LPD
Sbjct: 1851LMRTLTKSHEVLITPEEGSIGGFGSHVAXSLGVHALTDCELNTSPMLLPD 2000
>TC207142 similar to UP|Q8L692 (Q8L692) 1-deoxy-D-xylulose 5-phosphate
synthase 2 precursor, partial (55%)
Length = 1176
Score = 469 bits (1206), Expect = e-132
Identities = 237/389 (60%), Positives = 292/389 (74%), Gaps = 11/389 (2%)
Frame = +1
Query: 289 HELAAKVDEYARGMMGPPGSTLFEELGLYYIGPVDGHNIEDLISVLQEVASLDSMGPVLI 348
H++AAKVDEYARGM+ GSTLFEELGLYYIGPVDGHNIEDL+++ ++V ++ + GPVLI
Sbjct: 4 HQVAAKVDEYARGMISASGSTLFEELGLYYIGPVDGHNIEDLVTIFEKVKAMPAPGPVLI 183
Query: 349 HVITNENQ----VEEHNKKSYMTDKQQDDNAGRLQ------TYGDCFVESLVAEAEKDKD 398
HV+T + + E+ + + K +L+ +Y F ESL+ EAE D
Sbjct: 184 HVVTEKGKGYPPAEKAADRMHGVVKFDPKTGHQLKAKSSTLSYTQYFAESLIKEAEIDNK 363
Query: 399 IVVVHAGITTEPSLKLFMEKFPDRIFNVGIAEQHAVTFASGLSCGGLKPFCIIPSSFLQR 458
IV +HA + L F +KFP+R F+VGIAEQHAVTFA+GL+ GLKPFC I SSFLQR
Sbjct: 364 IVAIHAAMGGGTGLNYFQKKFPERCFDVGIAEQHAVTFAAGLAAEGLKPFCAIYSSFLQR 543
Query: 459 AYDQVVHDVDQQKVPVRFVITSAGLVGSDGPLQCGAFDITFMSCLPNMIVMAPSDEAELV 518
YDQVVHDVD QK+PVRF + AGLVG+DGP CGAFDIT+M+CLPNM+VMAPSDE EL+
Sbjct: 544 GYDQVVHDVDLQKLPVRFALDRAGLVGADGPTHCGAFDITYMACLPNMVVMAPSDETELM 723
Query: 519 HMVATAAHINDQPVCFRYPRGALVGKDEAILD-GIPIEIGKGRILVEGKDVALLGYGSMV 577
HMVATAA I+D+P CFR+PRG +G + + G P+EIGKGRILVEG VA+LGYGS+V
Sbjct: 724 HMVATAAAIDDRPSCFRFPRGNGIGATLPLNNKGTPLEIGKGRILVEGSRVAILGYGSVV 903
Query: 578 QNCLKAYSLLANLGIEVTVADARFCKPLDIELLRQLCKHHSFLITVEEGSIGGFGSHVAQ 637
Q C +A +L LGI+VTVADARFCKPLD L+R L K H LITVEEGSIGGFGSHV+Q
Sbjct: 904 QQCRQAS*MLKELGIDVTVADARFCKPLDTGLIRLLAKEHEILITVEEGSIGGFGSHVSQ 1083
Query: 638 FIALDGLLDRRIKWRPIVLPDSYIEHASP 666
F++L G+LD +KWR ++LPD YIEH SP
Sbjct: 1084FLSLSGILDGPLKWRAMMLPDRYIEHGSP 1170
>TC213419 similar to UP|Q9LFL9 (Q9LFL9) 1-D-deoxyxylulose 5-phosphate
synthase-like protein, partial (21%)
Length = 492
Score = 240 bits (612), Expect(2) = 6e-68
Identities = 118/138 (85%), Positives = 128/138 (92%)
Frame = +3
Query: 562 LVEGKDVALLGYGSMVQNCLKAYSLLANLGIEVTVADARFCKPLDIELLRQLCKHHSFLI 621
L++ K A LGYGSMVQNCLKA+SLLA LG+EVTVADARFCKPLDI LLRQLCKHHSFL+
Sbjct: 39 LLKVKMXAFLGYGSMVQNCLKAHSLLAKLGMEVTVADARFCKPLDIMLLRQLCKHHSFLV 218
Query: 622 TVEEGSIGGFGSHVAQFIALDGLLDRRIKWRPIVLPDSYIEHASPNQQLNQAGLTGHHIA 681
TVEEGSIGGFGSHVAQFIA++GLLD RIKWRPIVLPD YIEHASPN+QL+Q GL+GHHIA
Sbjct: 219 TVEEGSIGGFGSHVAQFIAVNGLLDGRIKWRPIVLPDRYIEHASPNEQLDQTGLSGHHIA 398
Query: 682 ATALSLLGRTREALSFMC 699
ATALSLLGRTREAL FMC
Sbjct: 399 ATALSLLGRTREALLFMC 452
Score = 37.0 bits (84), Expect(2) = 6e-68
Identities = 15/19 (78%), Positives = 19/19 (99%)
Frame = +2
Query: 550 DGIPIEIGKGRILVEGKDV 568
+GIPI+IG+GR+LVEGKDV
Sbjct: 2 EGIPIKIGRGRVLVEGKDV 58
>AI794684 similar to GP|8953400|emb 1-D-deoxyxylulose 5-phosphate
synthase-like protein {Arabidopsis thaliana}, partial
(18%)
Length = 461
Score = 246 bits (629), Expect = 2e-65
Identities = 117/139 (84%), Positives = 129/139 (92%)
Frame = +3
Query: 373 DNAGRLQTYGDCFVESLVAEAEKDKDIVVVHAGITTEPSLKLFMEKFPDRIFNVGIAEQH 432
DN QTYG+CFV +LVAEAEKDKDI+VVHAG+T EPSL+LF EKFPDR F+VG+AEQH
Sbjct: 45 DNPVWPQTYGNCFVATLVAEAEKDKDIIVVHAGLTMEPSLELFQEKFPDRFFDVGMAEQH 224
Query: 433 AVTFASGLSCGGLKPFCIIPSSFLQRAYDQVVHDVDQQKVPVRFVITSAGLVGSDGPLQC 492
AVTFASGL+CGGLKPFC+IP SFLQRAYDQVVHDVDQQ++PVRFVITSAGLVGSDGPL+C
Sbjct: 225 AVTFASGLACGGLKPFCVIPPSFLQRAYDQVVHDVDQQRIPVRFVITSAGLVGSDGPLRC 404
Query: 493 GAFDITFMSCLPNMIVMAP 511
GAFDI FMSCLPNMIVMAP
Sbjct: 405 GAFDINFMSCLPNMIVMAP 461
>TC226475 homologue to UP|Q8L693 (Q8L693) 1-deoxy-D-xylulose 5-phosphate
synthase 1 precursor, partial (27%)
Length = 757
Score = 211 bits (536), Expect(2) = 3e-65
Identities = 105/148 (70%), Positives = 120/148 (80%)
Frame = +3
Query: 551 GIPIEIGKGRILVEGKDVALLGYGSMVQNCLKAYSLLANLGIEVTVADARFCKPLDIELL 610
G P+EIGKGRIL+EG+ VALLGYGS VQNCL A SL+ G+ +TVADARFCKPLD L+
Sbjct: 141 GTPLEIGKGRILIEGERVALLGYGSAVQNCLAAASLVECHGLRLTVADARFCKPLDRSLI 320
Query: 611 RQLCKHHSFLITVEEGSIGGFGSHVAQFIALDGLLDRRIKWRPIVLPDSYIEHASPNQQL 670
R L K H LITVEEGSIGGFGSHVAQF+ALDGLLD ++KWRPIVLPD YI+H SP QL
Sbjct: 321 RSLAKSHEVLITVEEGSIGGFGSHVAQFMALDGLLDGKLKWRPIVLPDRYIDHGSPADQL 500
Query: 671 NQAGLTGHHIAATALSLLGRTREALSFM 698
+ AGLT HIAAT ++LG+TREAL M
Sbjct: 501 SLAGLTPSHIAATVFNVLGQTREALEVM 584
Score = 57.0 bits (136), Expect(2) = 3e-65
Identities = 25/34 (73%), Positives = 28/34 (81%)
Frame = +2
Query: 506 MIVMAPSDEAELVHMVATAAHINDQPVCFRYPRG 539
M+ PSDEA+L HMVATAA IND+P CFRYPRG
Sbjct: 2 MVXXXPSDEADLFHMVATAAAINDRPSCFRYPRG 103
>TC230963 similar to UP|O64904 (O64904) 1-deoxyxylulose-5-phosphate synthase,
partial (26%)
Length = 947
Score = 234 bits (597), Expect = 1e-61
Identities = 115/191 (60%), Positives = 143/191 (74%), Gaps = 4/191 (2%)
Frame = +1
Query: 501 SCLPNMIVMAPSDEAELVHMVATAAHINDQPVCFRYPRGALVGKDEAILD----GIPIEI 556
+CLPNM+VMAPSDE EL+HM+ATAA I+D+P CFRYPRG +G IL G P+E+
Sbjct: 1 ACLPNMVVMAPSDETELMHMIATAAAIDDRPSCFRYPRGNGIG---TILPPNNKGTPLEV 171
Query: 557 GKGRILVEGKDVALLGYGSMVQNCLKAYSLLANLGIEVTVADARFCKPLDIELLRQLCKH 616
GKGR+L EG VAL+GYG+MVQ+C++A +L GI TV DARFCKPLD +L+ +L +
Sbjct: 172 GKGRVLKEGSRVALVGYGTMVQSCMEAAKVLEAHGISTTVVDARFCKPLDGDLMTRLARE 351
Query: 617 HSFLITVEEGSIGGFGSHVAQFIALDGLLDRRIKWRPIVLPDSYIEHASPNQQLNQAGLT 676
H LITVEEGSIGGFGSHV+ F+ L+GLLD +KWR + LPD YI H S Q+ AGL+
Sbjct: 352 HEILITVEEGSIGGFGSHVSHFLGLNGLLDGNLKWRALTLPDRYINHGSQTDQIQMAGLS 531
Query: 677 GHHIAATALSL 687
+HIA TALSL
Sbjct: 532 SNHIAVTALSL 564
>CA800097 homologue to PIR|T09543|T09 deoxyxylulose synthase (EC 2.2.1.-)
TKT2 precursor chloroplast [validated] - pepper,
partial (20%)
Length = 437
Score = 187 bits (474), Expect = 2e-47
Identities = 91/141 (64%), Positives = 109/141 (76%)
Frame = +3
Query: 367 TDKQQDDNAGRLQTYGDCFVESLVAEAEKDKDIVVVHAGITTEPSLKLFMEKFPDRIFNV 426
T KQ NA Q+Y F E+L+AEAE DKDIV +HA + + LF+ +FP R F+V
Sbjct: 18 TGKQFKSNAAT-QSYTTYFAEALIAEAEADKDIVGIHAAMGGGTGMNLFLRRFPTRCFDV 194
Query: 427 GIAEQHAVTFASGLSCGGLKPFCIIPSSFLQRAYDQVVHDVDQQKVPVRFVITSAGLVGS 486
GIAEQHAVTFA+GL+C GLKPFC I SSF+QRAYDQVVHDVD QK+PVRF + AGLVG+
Sbjct: 195 GIAEQHAVTFAAGLACEGLKPFCAIYSSFMQRAYDQVVHDVDLQKLPVRFAMDRAGLVGA 374
Query: 487 DGPLQCGAFDITFMSCLPNMI 507
DGP CGAFD+TFM+CLPNM+
Sbjct: 375 DGPTHCGAFDVTFMACLPNMV 437
>TC207143 similar to UP|Q8L692 (Q8L692) 1-deoxy-D-xylulose 5-phosphate
synthase 2 precursor, partial (21%)
Length = 752
Score = 186 bits (472), Expect = 3e-47
Identities = 91/144 (63%), Positives = 110/144 (76%)
Frame = +1
Query: 554 IEIGKGRILVEGKDVALLGYGSMVQNCLKAYSLLANLGIEVTVADARFCKPLDIELLRQL 613
+EIGKGRILV G VA+LGYGS+VQ C +L LGI+VTVADARFCKPLD L+R L
Sbjct: 28 LEIGKGRILVXGSRVAILGYGSVVQQCRXXXEMLKELGIDVTVADARFCKPLDTGLIRLL 207
Query: 614 CKHHSFLITVEEGSIGGFGSHVAQFIALDGLLDRRIKWRPIVLPDSYIEHASPNQQLNQA 673
K H LITVEEGSIGGFGSHV+QF++L G+LD +KWR ++LPD YIEH SP Q+ +A
Sbjct: 208 AKEHEILITVEEGSIGGFGSHVSQFLSLSGILDGPLKWRAMMLPDRYIEHGSPQVQIEEA 387
Query: 674 GLTGHHIAATALSLLGRTREALSF 697
GL+ IAAT LSL+ R ++AL F
Sbjct: 388 GLSSKQIAATVLSLMERPKQALLF 459
>TC229279 homologue to UP|Q9XH50 (Q9XH50) 1-D-deoxyxylulose 5-phosphate
synthase, partial (15%)
Length = 1074
Score = 153 bits (387), Expect = 2e-37
Identities = 75/104 (72%), Positives = 84/104 (80%)
Frame = +1
Query: 595 TVADARFCKPLDIELLRQLCKHHSFLITVEEGSIGGFGSHVAQFIALDGLLDRRIKWRPI 654
TVADARFCKPLD L+R L + H LITVEEGSIGGFGSHV QF+ALDGLLD ++KWRPI
Sbjct: 4 TVADARFCKPLDRSLIRSLAQSHEVLITVEEGSIGGFGSHVVQFMALDGLLDGKLKWRPI 183
Query: 655 VLPDSYIEHASPNQQLNQAGLTGHHIAATALSLLGRTREALSFM 698
VLPD YI+H SP QL+ AGLT HIAAT +LLG+TREAL M
Sbjct: 184 VLPDCYIDHGSPVDQLSAAGLTPSHIAATVFNLLGQTREALEVM 315
>BE802429 similar to GP|5059160|gb| 1-D-deoxyxylulose 5-phosphate synthase
{Lycopersicon esculentum}, partial (21%)
Length = 481
Score = 120 bits (301), Expect = 2e-27
Identities = 70/151 (46%), Positives = 93/151 (61%), Gaps = 10/151 (6%)
Frame = +2
Query: 344 GPVLIHVITNENQVEEHNKKS---YMTDKQQDDNAGR-------LQTYGDCFVESLVAEA 393
G VLIHVIT + + +K+ Y D A + QTY F E+L+AE+
Sbjct: 29 GSVLIHVITEKCPGCPYAQKACHQYHGVTNFDPPAVKHFRSNATTQTYTTYFAEALIAES 208
Query: 394 EKDKDIVVVHAGITTEPSLKLFMEKFPDRIFNVGIAEQHAVTFASGLSCGGLKPFCIIPS 453
E DK V +HA + + LF +FP R F+VGIAEQHAVTFA+GL+ GLKP C I S
Sbjct: 209 EPDKHFVAIHAAMGGGTGMNLFHRRFPTRCFDVGIAEQHAVTFAAGLAREGLKPICAIYS 388
Query: 454 SFLQRAYDQVVHDVDQQKVPVRFVITSAGLV 484
SF+QR++DQVVHDVD K+ R+V+ AG++
Sbjct: 389 SFMQRSFDQVVHDVD*HKLTCRYVMDIAGII 481
>TC232096 similar to UP|DXS_CAPAN (O78328) Probable
1-deoxy-D-xylulose-5-phosphate synthase, chloroplast
precursor (1-deoxyxylulose-5-phosphate synthase) (DXP
synthase) (DXPS) (CapTKT2) , partial (13%)
Length = 1248
Score = 100 bits (250), Expect = 2e-21
Identities = 51/92 (55%), Positives = 63/92 (68%), Gaps = 1/92 (1%)
Frame = +3
Query: 466 DVDQQKVPVRFVITSAGLVGSDGPLQCGAFDITFMSCLPNMIVMAPSDEAELVHMVATAA 525
DVD Q + VRF + GLVG DG GAFD+TF +CLPNM+VM SD+AE+ H VATAA
Sbjct: 237 DVDVQHLLVRFAVDRGGLVGPDGSTHYGAFDVTFTACLPNMVVMVASDDAEIFHTVATAA 416
Query: 526 HINDQPVCFRYPRGALVGKD-EAILDGIPIEI 556
I+DQP CFRY +G VG + GIP+E+
Sbjct: 417 AISDQPCCFRYQKGNGVGVEIPPGNKGIPLEV 512
>TC223958 similar to UP|DXS_CAPAN (O78328) Probable
1-deoxy-D-xylulose-5-phosphate synthase, chloroplast
precursor (1-deoxyxylulose-5-phosphate synthase) (DXP
synthase) (DXPS) (CapTKT2) , partial (12%)
Length = 520
Score = 95.5 bits (236), Expect = 7e-20
Identities = 47/91 (51%), Positives = 62/91 (67%), Gaps = 1/91 (1%)
Frame = +3
Query: 467 VDQQKVPVRFVITSAGLVGSDGPLQCGAFDITFMSCLPNMIVMAPSDEAELVHMVATAAH 526
+ ++ + VRF + GLVG DG GAFD+TF +CLPNM+VM SD+AE+ H VATAA
Sbjct: 138 IGEKHLLVRFAVDRGGLVGPDGSTHYGAFDVTFTACLPNMVVMVASDDAEIFHTVATAAA 317
Query: 527 INDQPVCFRYPRGALVGKD-EAILDGIPIEI 556
I+DQP CFRY +G VG + GIP+E+
Sbjct: 318 ISDQPCCFRYQKGNGVGVEIPPGNKGIPLEV 410
>BM253046 homologue to GP|21322713|emb 1-deoxy-D-xylulose 5-phosphate
synthase 1 {Medicago truncatula}, partial (8%)
Length = 303
Score = 66.2 bits (160), Expect(2) = 2e-16
Identities = 31/41 (75%), Positives = 35/41 (84%)
Frame = -2
Query: 610 LRQLCKHHSFLITVEEGSIGGFGSHVAQFIALDGLLDRRIK 650
+R L K H LITVEEGSIGGFGSHVAQF+ALDGLLD ++K
Sbjct: 302 IRSLAKSHEVLITVEEGSIGGFGSHVAQFMALDGLLDGKLK 180
Score = 38.5 bits (88), Expect(2) = 2e-16
Identities = 15/23 (65%), Positives = 18/23 (78%)
Frame = -1
Query: 649 IKWRPIVLPDSYIEHASPNQQLN 671
++WRPIVLPD YI+H SP Q N
Sbjct: 69 MQWRPIVLPDRYIDHGSPADQSN 1
>TC215667 similar to UP|ODPB_PEA (P52904) Pyruvate dehydrogenase E1 component
beta subunit, mitochondrial precursor (PDHE1-B) ,
complete
Length = 1464
Score = 62.8 bits (151), Expect = 5e-10
Identities = 62/229 (27%), Positives = 97/229 (42%), Gaps = 15/229 (6%)
Frame = +1
Query: 413 KLFMEKF-PDRIFNVGIAEQHAVTFASGLSCGGLKPFC-IIPSSFLQRAYDQVVHDV--- 467
K ++K+ P+R+ + I E G + GL+P + +F +A D +++
Sbjct: 265 KGLLDKYGPERVLDTPITEAGFAGIGVGAAYYGLRPVVEFMTFNFSMQAIDHIINSAAKS 444
Query: 468 -----DQQKVPVRFVITSAGLVGSDGPLQCGAFDITFMSCLPNMIVMAPSDEAELVHMVA 522
Q VP+ F + G G + + SC P + V++P + ++
Sbjct: 445 NYMSAGQISVPIVFRGPNGAAAGV-GAQHSQCYASLYGSC-PGLKVLSPYSSEDARGLLK 618
Query: 523 TAAHINDQPVCFRYPRGALVGKD-----EAILDGIPIEIGKGRILVEGKDVALLGYGSMV 577
A D PV F L G+ E + + IGK +I EGKDV + Y MV
Sbjct: 619 AAIRDPD-PVVF-LENELLYGESFPVSAEVLDSSFCLPIGKAKIEREGKDVTITAYSKMV 792
Query: 578 QNCLKAYSLLANLGIEVTVADARFCKPLDIELLRQLCKHHSFLITVEEG 626
LKA LA GI V + R +PLD + + + L+TVEEG
Sbjct: 793 GYALKAAETLAKEGISAEVINLRSIRPLDRSTINASVRKTNRLVTVEEG 939
>TC225970 homologue to GB|AAB86804.1|2454184|ATU80186 pyruvate dehydrogenase
E1 beta subunit {Arabidopsis thaliana;} , partial (80%)
Length = 1579
Score = 47.8 bits (112), Expect = 2e-05
Identities = 65/270 (24%), Positives = 111/270 (41%), Gaps = 17/270 (6%)
Frame = +3
Query: 387 ESLVAEAEKDKDIVVV-----HAGITTEPSLKLFMEKFPD-RIFNVGIAEQHAVTFASGL 440
E L E E+D + V+ H G + + + K KF D R+ + IAE G
Sbjct: 435 EGLEEEMERDPCVCVMGEDVGHYGGSYKVT-KGLATKFGDLRVLDTPIAENSFTGMGIGA 611
Query: 441 SCGGLKPFCI-IPSSFLQRAYDQVVHDVD--------QQKVPVRFVITSAGLVGSDGPLQ 491
+ GL+P + FL A++Q+ ++ Q K+P+ VI G VG +
Sbjct: 612 AMTGLRPVVEGMNMGFLLLAFNQISNNCGMLHYTSGGQFKIPI--VIRGPGGVGRQLGAE 785
Query: 492 CGAFDITFMSCLPNMIVMAPSDEAELVHMVATAAHINDQPVCFRYPRGALVGKDEAILDG 551
++ +P + ++A S ++ A + + F + L E I D
Sbjct: 786 HSQRLESYFQSIPGIQMVACSTPYNAKGLMKAAIRSENPVILFEHV--LLYNLKERIPDE 959
Query: 552 -IPIEIGKGRILVEGKDVALLGYGSMVQNCLKAYSLLANLGIEVTVADARFCKPLDIELL 610
+ + + ++ G+ V +L Y M + ++A L N G + V D R KP D+ +
Sbjct: 960 EYVLSLEEAEMVRPGEHVTILTYSRMRYHVMQAAKTLVNKGYDPEVIDIRSLKPFDLHTI 1139
Query: 611 -RQLCKHHSFLITVEEGSIGGFGSHVAQFI 639
+ K H LI E GG G+ + I
Sbjct: 1140GNSVKKTHRVLIVEECMRTGGIGASLTAAI 1229
>AW156487 homologue to GP|21322713|emb 1-deoxy-D-xylulose 5-phosphate
synthase 1 {Medicago truncatula}, partial (9%)
Length = 293
Score = 33.1 bits (74), Expect(2) = 2e-04
Identities = 13/25 (52%), Positives = 19/25 (76%)
Frame = +2
Query: 57 DFYWEKVPTPILDTVQNPLCLKNLS 81
++Y + PTP+LDTV P+ +KNLS
Sbjct: 32 EYYSHRPPTPLLDTVNYPIHMKNLS 106
Score = 30.4 bits (67), Expect(2) = 2e-04
Identities = 18/39 (46%), Positives = 25/39 (63%)
Frame = +1
Query: 79 NLSQQELKQLAAEIRLELSSILSGTQILLNPSMAVVDLT 117
+L QELKQLA E+R ++ +S T L S+ VV+LT
Sbjct: 175 DLLMQELKQLADELRSDVIFSVSRTGGHLGSSLGVVELT 291
>TC225973 homologue to GB|AAB86804.1|2454184|ATU80186 pyruvate dehydrogenase
E1 beta subunit {Arabidopsis thaliana;} , partial (58%)
Length = 983
Score = 37.7 bits (86), Expect = 0.017
Identities = 24/87 (27%), Positives = 40/87 (45%), Gaps = 1/87 (1%)
Frame = +1
Query: 554 IEIGKGRILVEGKDVALLGYGSMVQNCLKAYSLLANLGIEVTVADARFCKPLDIELL-RQ 612
+ + + ++ G+ V +L Y M + ++A L N G + V D R KP D+ +
Sbjct: 418 LSLEEAEMVRPGEHVTILTYSRMRYHVMQAAKTLVNKGYDPEVIDIRSLKPFDLHTIGNS 597
Query: 613 LCKHHSFLITVEEGSIGGFGSHVAQFI 639
+ K H LI E GG G+ + I
Sbjct: 598 VKKTHRVLIVEECMRTGGIGASLTAAI 678
>TC226110 similar to UP|TKTC_CRAPL (Q42676) Transketolase, chloroplast (TK)
(Fragment) , partial (90%)
Length = 1719
Score = 29.6 bits (65), Expect = 4.7
Identities = 31/130 (23%), Positives = 50/130 (37%), Gaps = 7/130 (5%)
Frame = +2
Query: 474 VRFVITSAGL-VGSDGPLQCGAFDITFMSCLPNMIVMAPSDEAELVHMVATAAHINDQPV 532
V +V+T + +G DGP + +PN++++ P+D E A +P
Sbjct: 770 VIYVMTHDSIGLGEDGPTHQPIEHLASFRAMPNILMLRPADGNETAGAYKVAVLNRKRPS 949
Query: 533 CFRYPRGALVGKDEAILDGIPIEIGKGRILVEGK------DVALLGYGSMVQNCLKAYSL 586
R L ++G+ KG + DV L+G GS ++ KA
Sbjct: 950 ILALSRQKLPQLPGTSIEGVE----KGGYTISDNSTGNKPDVILIGTGSELEIAAKAADD 1117
Query: 587 LANLGIEVTV 596
L G V V
Sbjct: 1118LRKEGKAVRV 1147
>TC221597
Length = 838
Score = 28.9 bits (63), Expect = 8.1
Identities = 20/71 (28%), Positives = 37/71 (51%), Gaps = 3/71 (4%)
Frame = -2
Query: 441 SCGGLKPFCIIPSSFLQRAYDQVVHDVDQQKVPVRFVITSAG---LVGSDGPLQCGAFDI 497
+CG FCI+P F QR+ ++ +V+ +++ + +T G L G+D AF+
Sbjct: 621 NCGFFP*FCILPVKFGQRSL-LILEEVEGKRIYCFYCVTEEGWFLLQGTDSL----AFNK 457
Query: 498 TFMSCLPNMIV 508
+ C N++V
Sbjct: 456 ATILCRSNILV 424
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.137 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,314,892
Number of Sequences: 63676
Number of extensions: 395588
Number of successful extensions: 1712
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 1704
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1706
length of query: 700
length of database: 12,639,632
effective HSP length: 104
effective length of query: 596
effective length of database: 6,017,328
effective search space: 3586327488
effective search space used: 3586327488
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC148176.1


