

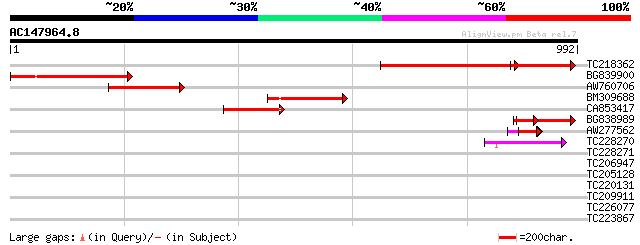
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147964.8 + phase: 0
(992 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC218362 similar to UP|Q8W338 (Q8W338) TGF beta receptor associa... 353 3e-97
BG839900 326 4e-89
AW760706 199 7e-51
BM309688 197 1e-50
CA853417 153 3e-37
BG838989 100 5e-21
AW277562 64 3e-10
TC228270 55 2e-07
TC228271 37 0.043
TC206947 similar to UP|O04470 (O04470) F5I14.6 protein, partial ... 33 0.62
TC205128 similar to UP|Q9SMA5 (Q9SMA5) Zwh0004.1, partial (8%) 32 1.8
TC220131 similar to UP|ATK2_ARATH (P46864) Kinesin 2 (Kinesin-li... 30 4.0
TC209911 weakly similar to UP|Q94BZ1 (Q94BZ1) AT5g13750/MXE10_2 ... 30 5.3
TC226077 similar to UP|Q94C38 (Q94C38) AT3g42050/F4M19_10, parti... 30 6.9
TC223867 similar to GB|AAL60196.1|18139887|AF441079 O-linked N-a... 29 9.0
>TC218362 similar to UP|Q8W338 (Q8W338) TGF beta receptor associated
protein-like protein, partial (23%)
Length = 1473
Score = 353 bits (905), Expect = 3e-97
Identities = 184/245 (75%), Positives = 207/245 (84%), Gaps = 1/245 (0%)
Frame = +1
Query: 649 SMNEGAVSGTLQNEMVHLYLSEVLDWHADLSSEQKWDEKVYSPKRKKLLSALESISGYNP 708
+MNE AVSG LQNEMVH+YLSEVLDWHADLS++QKWDEK +SP RKKLL+ALESI+GYNP
Sbjct: 1 AMNENAVSGNLQNEMVHIYLSEVLDWHADLSAQQKWDEKDHSPTRKKLLTALESIAGYNP 180
Query: 709 EALLKLLPSDALYEERAILLGKMNQHELALSLYVHKLHVPELALSYCDHVYESAHKSSVK 768
EALLK LP DALYEE AILLGKMN+HELALSLYV KL+ PELALSYCD VYES H+ S K
Sbjct: 181 EALLKRLPPDALYEEHAILLGKMNRHELALSLYVLKLNAPELALSYCDRVYESMHQPSAK 360
Query: 769 SLSNIYLMLLQIYLNPRRTTKNYEKKISNLLSPRNKSIRKVTSKSLSRTMSRGSKKIAAI 828
+ SNIYL+LLQIYLNPRRTT +E +I+NLLSP+NK+I K+T ++ RGSKKIAAI
Sbjct: 361 NSSNIYLVLLQIYLNPRRTTAGFENRITNLLSPQNKTIPKLTPTPSIKSRGRGSKKIAAI 540
Query: 829 EIAEDAKASQSS-DSGRSDADTEEFTEEECTSIMLDEALDLLSRRWDRINGAQALKLLPK 887
E AED K S SS DSGRSD D +E+ + T IMLDE LDLLSRRWDRINGAQALKLLPK
Sbjct: 541 EGAEDTKVSLSSTDSGRSDGDADEYNDGSPT-IMLDEILDLLSRRWDRINGAQALKLLPK 717
Query: 888 ETKLQ 892
ETKLQ
Sbjct: 718 ETKLQ 732
Score = 177 bits (449), Expect = 2e-44
Identities = 89/115 (77%), Positives = 102/115 (88%), Gaps = 1/115 (0%)
Frame = +1
Query: 877 NGAQALKLLPK-ETKLQNLLPILGPLVRKSSEMYRNCSVVRSLRQSENLQVKDELYNKRK 935
+GAQALKL K + LQ+LL LGPL+RKSSEMYRNCSV++SLRQSENLQVKDELY++RK
Sbjct: 862 HGAQALKLGNKRDNNLQDLLSFLGPLLRKSSEMYRNCSVIKSLRQSENLQVKDELYSQRK 1041
Query: 936 AVIKISDDNMCSLCHKKIGTSVFAVYPNGKTLVHFVCFRDSQSMKAVAKVSPLKK 990
V+KI+ D+MCSLCHKKIGTSVFAVYPNG TLVHFVCFRDSQ+MKAV K S L+K
Sbjct: 1042EVVKITGDSMCSLCHKKIGTSVFAVYPNGSTLVHFVCFRDSQNMKAVGKGSQLRK 1206
>BG839900
Length = 716
Score = 326 bits (835), Expect = 4e-89
Identities = 161/215 (74%), Positives = 184/215 (84%)
Frame = +1
Query: 1 MVHTAYDSFELLTNSTSKIESIESYGSKLLLGCSNGSLLVYAPEHSVPAPEEMRKEAYVL 60
MVH+AYD EL+ +KIESIESY SKLL+GCS+GSL ++APE + ++Y L
Sbjct: 52 MVHSAYDCLELVRECPAKIESIESYDSKLLVGCSDGSLRIFAPE--TESSSSNGSKSYAL 225
Query: 61 ERNVNGFAKKAVVSLQVVESREFLLSLSESIAFHKLPTFETIAVITKAKGANAFCWDEHR 120
E+N+ GFAKK+V+S+ VVESREFL+SLSESIAFH+LP+FETIAVITKAKGAN FCWD R
Sbjct: 226 EKNLAGFAKKSVLSMAVVESREFLISLSESIAFHRLPSFETIAVITKAKGANVFCWDHRR 405
Query: 121 GFLCFARQKRVCIFRRDGGRGFVEVKDFGVLDVVKSMSWCGENICLGIRKAYVILNATSG 180
GFLCFARQKRVCIFR DGGRGFVEVKDFGV D VKSM WCGENICLGIR+ YVILNAT+G
Sbjct: 406 GFLCFARQKRVCIFRHDGGRGFVEVKDFGVADTVKSMCWCGENICLGIRREYVILNATNG 585
Query: 181 SISEVFTSGRLAPPLVVSLPSGELLLGKDNIGVIV 215
++SEVFTSGRLAPPLV SLPSGELLLGK+NIGV V
Sbjct: 586 ALSEVFTSGRLAPPLVXSLPSGELLLGKENIGVXV 690
>AW760706
Length = 397
Score = 199 bits (505), Expect = 7e-51
Identities = 101/132 (76%), Positives = 111/132 (83%)
Frame = +2
Query: 174 ILNATSGSISEVFTSGRLAPPLVVSLPSGELLLGKDNIGVIVDQNGKLRPEGRICWSEAP 233
ILNAT+G++SEVFT GRL PPLVV PSGEL LGK+NIGV VDQNGKL PE RICWS P
Sbjct: 2 ILNATNGALSEVFTFGRLTPPLVVYFPSGELFLGKENIGVFVDQNGKLLPERRICWS*TP 181
Query: 234 TEVVIQNPYALALLPRFVEIRSLRGPYPLIQTIVFRNVRHLRQSNNSVIIALENSIHCLF 293
EVVI PYA+AL PRF EIRSLR PY LIQT+V RNVRHL QSN+S I+AL+NSIH L+
Sbjct: 182 LEVVIHKPYAIAL*PRFEEIRSLRAPYTLIQTVVLRNVRHLCQSNDSGILALDNSIHGLY 361
Query: 294 PVPLGAQIVQLT 305
PVPLGAQIVQLT
Sbjct: 362 PVPLGAQIVQLT 397
>BM309688
Length = 419
Score = 197 bits (502), Expect = 1e-50
Identities = 101/141 (71%), Positives = 122/141 (85%)
Frame = +2
Query: 451 ATAEGTEEVVFDAVGNNFESYNSNRFKKINKRHGSIPVSSEAREMASILDTALLQAMLLT 510
ATAEGTEEVVFDAVG+NF SYN R KK NK G++PVSS AREMAS+LDTALL+A+LLT
Sbjct: 2 ATAEGTEEVVFDAVGDNFASYN--RLKKTNKGRGNVPVSSGAREMASMLDTALLEALLLT 175
Query: 511 GQPSMAENLLRVLNYCDLKICEEILQEGSYHVSLVELYKCNSMHREALEIINKSVKESES 570
GQ S+A LLR +NYCDLKICEEIL++G++HV+L+ELYK NS+HREALE+++K V E +S
Sbjct: 176 GQSSVALELLRGVNYCDLKICEEILRKGNHHVALLELYKHNSLHREALELLHKLVDELKS 355
Query: 571 SQSKIAHRFKPEAIIEYLKPL 591
SQS+I RFKPE I+EYLKPL
Sbjct: 356 SQSEITQRFKPEDIVEYLKPL 418
>CA853417
Length = 580
Score = 153 bits (387), Expect = 3e-37
Identities = 82/109 (75%), Positives = 92/109 (84%), Gaps = 1/109 (0%)
Frame = +3
Query: 374 ILPKTTIVHDSDKLDIFGDPLHLSRGSS-MSDDMEPSSASNMSELDDNAELESKKMSHNM 432
ILPKTTIV+D +KLDI+GD +LSR SS +SDDMEPSS S+M E D+NA LESKKM+HNM
Sbjct: 6 ILPKTTIVYDPEKLDIYGDASYLSRASSGVSDDMEPSSTSHMPESDENAALESKKMNHNM 185
Query: 433 LMALIKFLHKKRHSIIEKATAEGTEEVVFDAVGNNFESYNSNRFKKINK 481
LMALIK+L KKR S IEKATAEGTEEVVFDAVG+NF SY NR KK NK
Sbjct: 186 LMALIKYLQKKRFSFIEKATAEGTEEVVFDAVGDNFASY--NRLKKTNK 326
>BG838989
Length = 689
Score = 99.8 bits (247), Expect = 5e-21
Identities = 58/107 (54%), Positives = 68/107 (63%), Gaps = 3/107 (2%)
Frame = +2
Query: 887 KETKLQNLLPILGPLVR---KSSEMYRNCSVVRSLRQSENLQVKDELYNKRKAVIKISDD 943
K Q+LL LGPL K +E+ R V +R Q KD+ Y+ RKAV+KI+ D
Sbjct: 17 KGRNYQDLLXFLGPLXENLVKCTEIVR**RV*DKVR---TFQGKDKXYSXRKAVVKITGD 187
Query: 944 NMCSLCHKKIGTSVFAVYPNGKTLVHFVCFRDSQSMKAVAKVSPLKK 990
+MC LCHKKIGTSVFAVYPNG V FVCFRD Q+MKAV K L K
Sbjct: 188 SMCFLCHKKIGTSVFAVYPNGSXXVXFVCFRDXQNMKAVGKGFQLXK 328
Score = 51.6 bits (122), Expect = 2e-06
Identities = 26/43 (60%), Positives = 29/43 (66%)
Frame = +1
Query: 882 LKLLPKETKLQNLLPILGPLVRKSSEMYRNCSVVRSLRQSENL 924
L PK TKL + G RK SEMYRNCSV++SLRQSENL
Sbjct: 1 LNFYPKGTKLPGSTXVSGTPXRKFSEMYRNCSVIKSLRQSENL 129
>AW277562
Length = 431
Score = 63.9 bits (154), Expect = 3e-10
Identities = 31/40 (77%), Positives = 35/40 (87%)
Frame = +2
Query: 891 LQNLLPILGPLVRKSSEMYRNCSVVRSLRQSENLQVKDEL 930
LQ+LL LGPL+RKSSEMYRNCSV++SLRQSENLQV L
Sbjct: 227 LQDLLSFLGPLLRKSSEMYRNCSVIKSLRQSENLQVSTYL 346
Score = 52.0 bits (123), Expect = 1e-06
Identities = 30/60 (50%), Positives = 35/60 (58%)
Frame = +3
Query: 872 RWDRINGAQALKLLPKETKLQNLLPILGPLVRKSSEMYRNCSVVRSLRQSENLQVKDELY 931
RWDRINGAQALKLLPKETKLQ G + +K Y NC + E V+ +LY
Sbjct: 3 RWDRINGAQALKLLPKETKLQVSTTCWGTIAKK----YTNCFYWSNFCS*EFCSVRRKLY 170
>TC228270
Length = 1186
Score = 54.7 bits (130), Expect = 2e-07
Identities = 36/149 (24%), Positives = 76/149 (50%), Gaps = 6/149 (4%)
Frame = +3
Query: 832 EDAKASQS--SDSGRSDADTE----EFTEEECTSIMLDEALDLLSRRWDRINGAQALKLL 885
ED++A++ ++ GR+DA + ++ M A+ LL + ++ Q L+ L
Sbjct: 183 EDSEAAEQYCAEIGRADAYMQLLEMYLDPQDDKDPMFTAAVRLLHNHGESLDPLQVLEKL 362
Query: 886 PKETKLQNLLPILGPLVRKSSEMYRNCSVVRSLRQSENLQVKDELYNKRKAVIKISDDNM 945
+ LQ L + R +R +V +L ++ ++ + +R ++I+D+++
Sbjct: 363 SPDMPLQLASDTLLRMFRARVHHHRQGQIVHNLSRAVDIDARLSRLEERSRNVQINDESL 542
Query: 946 CSLCHKKIGTSVFAVYPNGKTLVHFVCFR 974
C C ++GT +FA+YP+ T+V + C+R
Sbjct: 543 CDSCDARLGTKLFAMYPD-DTVVCYKCYR 626
>TC228271
Length = 609
Score = 37.0 bits (84), Expect = 0.043
Identities = 13/37 (35%), Positives = 28/37 (75%)
Frame = +2
Query: 938 IKISDDNMCSLCHKKIGTSVFAVYPNGKTLVHFVCFR 974
++I+D+++C C ++GT +FA+YP+ ++V + C+R
Sbjct: 17 VQINDESLCDSCDARLGTKLFAMYPD-DSVVCYKCYR 124
>TC206947 similar to UP|O04470 (O04470) F5I14.6 protein, partial (53%)
Length = 1553
Score = 33.1 bits (74), Expect = 0.62
Identities = 27/103 (26%), Positives = 52/103 (50%)
Frame = +2
Query: 498 ILDTALLQAMLLTGQPSMAENLLRVLNYCDLKICEEILQEGSYHVSLVELYKCNSMHREA 557
++DT +Q L + S++E R L + +++ EE+++E +V+ NS ++
Sbjct: 509 VVDT--IQVTSLPSEDSVSERR-RKLEFLEMQ--EELIKEEEEREEVVQARMENSSSQD- 670
Query: 558 LEIINKSVKESESSQSKIAHRFKPEAIIEYLKPLCELDTTLVL 600
+K++KE S +K AH+ + E + LCEL L +
Sbjct: 671 ----DKALKEMNISTAKEAHQLARDRAFENKEQLCELSRALAV 787
>TC205128 similar to UP|Q9SMA5 (Q9SMA5) Zwh0004.1, partial (8%)
Length = 411
Score = 31.6 bits (70), Expect = 1.8
Identities = 26/93 (27%), Positives = 45/93 (47%)
Frame = +1
Query: 321 PPEDSNLRAAKEDSIHIRYAHYLFDNGSYEESMEHFLASQVDITYVLSLYTSIILPKTTI 380
PP SN K D H+ + H+L +S + ++ +T LS + + +PK+ I
Sbjct: 136 PPSISN----KLD--HVDHHHHLITLRPTTKSSYNSSEAEPSLTLGLSRESYLKVPKSII 297
Query: 381 VHDSDKLDIFGDPLHLSRGSSMSDDMEPSSASN 413
H+++K+ DPL LS ++ SS S+
Sbjct: 298 GHNNNKVSSCDDPLDLSTQTNSPHHSAVSSFSS 396
>TC220131 similar to UP|ATK2_ARATH (P46864) Kinesin 2 (Kinesin-like protein
B), partial (37%)
Length = 1044
Score = 30.4 bits (67), Expect = 4.0
Identities = 12/19 (63%), Positives = 16/19 (84%)
Frame = +1
Query: 427 KMSHNMLMALIKFLHKKRH 445
KM+ NML+ LIKFLH++ H
Sbjct: 298 KMAKNMLLHLIKFLHQRHH 354
>TC209911 weakly similar to UP|Q94BZ1 (Q94BZ1) AT5g13750/MXE10_2
(Transporter-like protein), partial (44%)
Length = 925
Score = 30.0 bits (66), Expect = 5.3
Identities = 17/49 (34%), Positives = 27/49 (54%), Gaps = 1/49 (2%)
Frame = +1
Query: 342 YLFDNGSYEESMEHFLASQVDITYVLSLYTSII-LPKTTIVHDSDKLDI 389
YLF GS+ + +FL V T+ S+ + I LP+T H+ + L+I
Sbjct: 598 YLFSKGSFWDRFPYFLPCLVISTFAFSVAIACIWLPETNHNHNPEALEI 744
>TC226077 similar to UP|Q94C38 (Q94C38) AT3g42050/F4M19_10, partial (44%)
Length = 1035
Score = 29.6 bits (65), Expect = 6.9
Identities = 25/82 (30%), Positives = 34/82 (40%), Gaps = 10/82 (12%)
Frame = +3
Query: 617 LFLSGNIPADMVNLYLKQHAPNLQAT------YLELVLSMNEGAVSGTLQNEMVHLYLSE 670
L G + A MV+L L Q +L+A LE + S+ EG + +Y E
Sbjct: 69 LMSKGTLGAQMVDLQLAQVVQSLKAQAWSDEDLLEALNSLEEGLKDNIKRLSSFDMYKQE 248
Query: 671 VLDWHADLSSEQK----WDEKV 688
VL H D S K W E +
Sbjct: 249 VLLGHLDWSPMHKDPIFWRENI 314
>TC223867 similar to GB|AAL60196.1|18139887|AF441079 O-linked N-acetyl
glucosamine transferase {Arabidopsis thaliana;} ,
partial (8%)
Length = 591
Score = 29.3 bits (64), Expect = 9.0
Identities = 12/37 (32%), Positives = 22/37 (59%)
Frame = +3
Query: 319 LLPPEDSNLRAAKEDSIHIRYAHYLFDNGSYEESMEH 355
LLP + ED +H+ AH ++ G+Y++++EH
Sbjct: 333 LLPLRGHDSSEVDED-VHLSLAHQMYKTGNYKQALEH 440
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.133 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 40,974,218
Number of Sequences: 63676
Number of extensions: 563596
Number of successful extensions: 2937
Number of sequences better than 10.0: 31
Number of HSP's better than 10.0 without gapping: 2910
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2932
length of query: 992
length of database: 12,639,632
effective HSP length: 106
effective length of query: 886
effective length of database: 5,889,976
effective search space: 5218518736
effective search space used: 5218518736
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Medicago: description of AC147964.8


