

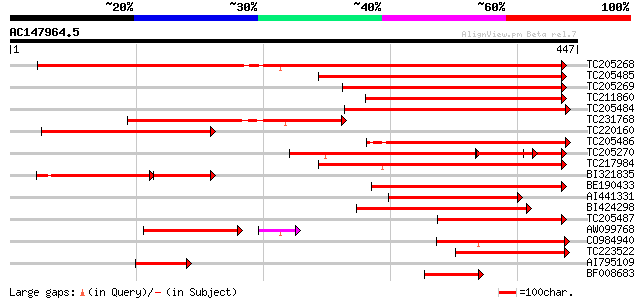
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147964.5 + phase: 0
(447 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC205268 similar to UP|Q6Z001 (Q6Z001) Membrane protein-like, pa... 534 e-152
TC205485 similar to GB|AAO63445.1|28951043|BT005381 At2g25737 {A... 248 3e-66
TC205269 similar to UP|Q6Z001 (Q6Z001) Membrane protein-like, pa... 247 9e-66
TC211860 similar to GB|AAO63445.1|28951043|BT005381 At2g25737 {A... 246 1e-65
TC205484 similar to GB|AAO63445.1|28951043|BT005381 At2g25737 {A... 239 1e-63
TC231768 similar to UP|Q6Z001 (Q6Z001) Membrane protein-like, pa... 236 2e-62
TC220160 similar to UP|Q6Z001 (Q6Z001) Membrane protein-like, pa... 233 2e-61
TC205486 similar to GB|AAO63445.1|28951043|BT005381 At2g25737 {A... 221 4e-58
TC205270 similar to UP|Q6Z001 (Q6Z001) Membrane protein-like, pa... 178 1e-57
TC217984 similar to UP|Q8S9J0 (Q8S9J0) At2g36630/F1O11.26, parti... 189 2e-48
BI321835 128 4e-47
BE190433 164 1e-40
AI441331 159 2e-39
BI424298 150 9e-37
TC205487 similar to GB|AAO63445.1|28951043|BT005381 At2g25737 {A... 150 1e-36
AW099768 138 2e-35
CO984940 126 2e-29
TC223522 weakly similar to UP|Q6Z001 (Q6Z001) Membrane protein-l... 122 2e-28
AI795109 69 4e-12
BF008683 66 3e-11
>TC205268 similar to UP|Q6Z001 (Q6Z001) Membrane protein-like, partial (74%)
Length = 1678
Score = 534 bits (1376), Expect = e-152
Identities = 259/428 (60%), Positives = 334/428 (77%), Gaps = 11/428 (2%)
Frame = +1
Query: 23 KKTSFFWYSGESSYERVWPEMKFGWRIVVGSIVGFFGAALGSVGGVGGGGIFIPMLTLII 82
K +F W S S Y+ WP+++FGWRI+ G+I+GF G+A G+VGGVGGGGIF+ ML+LII
Sbjct: 169 KVVNFLWSSSGSGYQHTWPDIEFGWRIITGTIIGFLGSAFGTVGGVGGGGIFVTMLSLII 348
Query: 83 GFDPKSSTALSKCMITGAAGSTVYYNLRLRHPTLDMPLIDYDLALLFQPMLMLGISIGVA 142
GFD KS+TA+SKCMITG A +TV+YNL+ +HPTLDMP+IDYDLALLFQP+L+LGISIGVA
Sbjct: 349 GFDAKSATAISKCMITGGAAATVFYNLKQKHPTLDMPVIDYDLALLFQPVLVLGISIGVA 528
Query: 143 FNVMFADWMVTILLIILFIGTSTKALVKGIDTWKKETIMKKEAFEEAAQMLESGSTPDYA 202
FNV+FADWM+T+LL+I+F+G +TKA +KG++TWKKETI+KKE A Q +G+ + +
Sbjct: 529 FNVIFADWMITVLLLIIFVGIATKAFLKGVETWKKETIIKKET---ARQSQFNGT--ERS 693
Query: 203 SEEDYKSLPA----------DLQDEEVPLLDNIHWKELSVLMYVWVAFLIVQILKTYSKT 252
E Y+ LP E +L+N+ WK L VL VWV L +I K+++ T
Sbjct: 694 EEVAYEPLPGGPNTSNHKEPKKSKETGSILENVRWKALGVLFTVWVLILASEIAKSHTTT 873
Query: 253 CSIEYWLLNSLQVPIAISVTLFEAICLCKGTRVIASRGKEIT-WKFHKICLYCFCGIIAG 311
CS+EYW+LN LQVP+A+ T ++A+ L G RVIAS+G + T W+ H++ +YC CGI AG
Sbjct: 874 CSVEYWVLNLLQVPVALGATSYQAVLLYTGKRVIASKGDQRTQWRAHQLIMYCSCGICAG 1053
Query: 312 MVSGLLGLGGGFILGPLFLELGIPPQVASATSTFAMVFSSSMSVVQYYHLDRFPIPYASY 371
+V GLLGLGGGFILGPLFLELGIPPQV+SAT+TFAM FS+SMSVV+YY L RFPIPY Y
Sbjct: 1054IVGGLLGLGGGFILGPLFLELGIPPQVSSATATFAMTFSASMSVVEYYLLKRFPIPYTLY 1233
Query: 372 LVLVATIAALTGQHVVRKIIAIFGRASIIVFILAFTIFVSAISLGGVGIGNMVEKMENEE 431
V V+T AA GQ +VRK++AI GRAS+I+FIL+ TIFVSAISLGGVGI NM++++ N+E
Sbjct: 1234FVAVSTFAAFVGQVLVRKLVAILGRASLIIFILSSTIFVSAISLGGVGISNMIQRIANKE 1413
Query: 432 YMGFDNLC 439
YMGF+NLC
Sbjct: 1414YMGFENLC 1437
>TC205485 similar to GB|AAO63445.1|28951043|BT005381 At2g25737 {Arabidopsis
thaliana;} , partial (40%)
Length = 802
Score = 248 bits (634), Expect = 3e-66
Identities = 119/197 (60%), Positives = 156/197 (78%), Gaps = 1/197 (0%)
Frame = +1
Query: 244 QILKTYSKTCSIEYWLLNSLQVPIAISVTLFEAICLCKGTRVIASRGKE-ITWKFHKICL 302
QI + CS YW+LN LQ+PI++ V+ +EA L KG R I+S G + T+ ++ +
Sbjct: 1 QIAMNQTSKCSTIYWVLNMLQIPISVGVSGYEAASLYKGRRQISSVGDQGKTFTSQQLTI 180
Query: 303 YCFCGIIAGMVSGLLGLGGGFILGPLFLELGIPPQVASATSTFAMVFSSSMSVVQYYHLD 362
YC G++AG+V GLLG+GGGF++GPLFLELG+PPQV+SAT+TFAM FSSSMSVV+YY L
Sbjct: 181 YCIFGVLAGIVGGLLGIGGGFVMGPLFLELGVPPQVSSATATFAMTFSSSMSVVEYYLLK 360
Query: 363 RFPIPYASYLVLVATIAALTGQHVVRKIIAIFGRASIIVFILAFTIFVSAISLGGVGIGN 422
RFP+PYA Y + VA I+A+ GQH+VRK+I + GRAS+I+F+LAFTI VSA+SLGGVGI
Sbjct: 361 RFPVPYALYFIAVAAISAIVGQHIVRKLIDVLGRASLIIFVLAFTILVSAVSLGGVGIVA 540
Query: 423 MVEKMENEEYMGFDNLC 439
MV+K+EN EYMGFD+LC
Sbjct: 541 MVKKIENHEYMGFDDLC 591
>TC205269 similar to UP|Q6Z001 (Q6Z001) Membrane protein-like, partial (35%)
Length = 771
Score = 247 bits (630), Expect = 9e-66
Identities = 121/178 (67%), Positives = 148/178 (82%), Gaps = 1/178 (0%)
Frame = +3
Query: 263 LQVPIAISVTLFEAICLCKGTRVIASRGKEIT-WKFHKICLYCFCGIIAGMVSGLLGLGG 321
LQVP+A+ T ++A+ L G RVIAS+G + T W+ H++ LYC CGI AG+V GLLGLGG
Sbjct: 6 LQVPVALGATSYQAVLLYTGKRVIASKGDQRTQWRAHQLVLYCSCGICAGIVGGLLGLGG 185
Query: 322 GFILGPLFLELGIPPQVASATSTFAMVFSSSMSVVQYYHLDRFPIPYASYLVLVATIAAL 381
GFILGPLFLELGIPPQV+SAT+TFAM FS+SMSVV+YY L RFPIPY Y V V+T AA
Sbjct: 186 GFILGPLFLELGIPPQVSSATATFAMTFSASMSVVEYYLLKRFPIPYTLYFVAVSTFAAF 365
Query: 382 TGQHVVRKIIAIFGRASIIVFILAFTIFVSAISLGGVGIGNMVEKMENEEYMGFDNLC 439
GQ +VRK++AI GRAS+I+FIL+ TIFVSAISLGGVGI NM++K+ N+EYMGF+NLC
Sbjct: 366 VGQVLVRKLVAILGRASLIIFILSGTIFVSAISLGGVGISNMIQKIANKEYMGFENLC 539
>TC211860 similar to GB|AAO63445.1|28951043|BT005381 At2g25737 {Arabidopsis
thaliana;} , partial (33%)
Length = 626
Score = 246 bits (628), Expect = 1e-65
Identities = 124/160 (77%), Positives = 138/160 (85%), Gaps = 1/160 (0%)
Frame = +1
Query: 281 KGTRVIASRGKEIT-WKFHKICLYCFCGIIAGMVSGLLGLGGGFILGPLFLELGIPPQVA 339
K RV AS+G E+ K H+I YC GI+AGMV GLLGLGGGFILGPLFLELGIPPQVA
Sbjct: 1 KXXRVXASKGXEVXNXKIHQIXXYCSTGIMAGMVGGLLGLGGGFILGPLFLELGIPPQVA 180
Query: 340 SATSTFAMVFSSSMSVVQYYHLDRFPIPYASYLVLVATIAALTGQHVVRKIIAIFGRASI 399
SATSTFAMVFSSSMSVVQYY LDRFP+PYASY LVATIAA TGQHVVRK+I + GRASI
Sbjct: 181 SATSTFAMVFSSSMSVVQYYLLDRFPVPYASYFALVATIAAFTGQHVVRKVIVVLGRASI 360
Query: 400 IVFILAFTIFVSAISLGGVGIGNMVEKMENEEYMGFDNLC 439
I+FILA TIF+SAISLGGVGI N++EK+EN EYMGF++LC
Sbjct: 361 IIFILALTIFISAISLGGVGIENIIEKIENHEYMGFEDLC 480
>TC205484 similar to GB|AAO63445.1|28951043|BT005381 At2g25737 {Arabidopsis
thaliana;} , partial (37%)
Length = 821
Score = 239 bits (611), Expect = 1e-63
Identities = 114/179 (63%), Positives = 148/179 (81%), Gaps = 1/179 (0%)
Frame = +3
Query: 265 VPIAISVTLFEAICLCKGTRVIASRGKE-ITWKFHKICLYCFCGIIAGMVSGLLGLGGGF 323
VP+++ VT +EA L G RVIAS G++ + ++ +YC G++AG+V G+LGLGGGF
Sbjct: 3 VPVSVGVTAYEAAALFSGRRVIASTGEQGKDFTVLQLMIYCVFGVLAGVVGGMLGLGGGF 182
Query: 324 ILGPLFLELGIPPQVASATSTFAMVFSSSMSVVQYYHLDRFPIPYASYLVLVATIAALTG 383
++GPLFLELG+PPQV+SAT+TFAM FSSSMSV++YY L RFP+PYA Y +LVATIAA G
Sbjct: 183 VMGPLFLELGVPPQVSSATATFAMTFSSSMSVIEYYLLKRFPVPYALYFILVATIAAFVG 362
Query: 384 QHVVRKIIAIFGRASIIVFILAFTIFVSAISLGGVGIGNMVEKMENEEYMGFDNLCHHG 442
QH+VRK+I +FGRAS+I+FILA TIFVSA+SLGGVGI NMV K++N EYMGF++LC +G
Sbjct: 363 QHIVRKLIILFGRASLIIFILASTIFVSAVSLGGVGIVNMVHKIQNHEYMGFEDLCKYG 539
>TC231768 similar to UP|Q6Z001 (Q6Z001) Membrane protein-like, partial (21%)
Length = 504
Score = 236 bits (602), Expect = 2e-62
Identities = 125/175 (71%), Positives = 141/175 (80%), Gaps = 3/175 (1%)
Frame = +2
Query: 94 KCMITGAAGSTVYYNLRLRHPTLDMPLIDYDLALLFQPMLMLGISIGVAFNVMFADWMVT 153
KCMI GA+ STVYYNLRLRHPTLDMPLIDYDLAL+FQPMLMLGISIGV NVMFADWMVT
Sbjct: 2 KCMIMGASISTVYYNLRLRHPTLDMPLIDYDLALIFQPMLMLGISIGVICNVMFADWMVT 181
Query: 154 ILLIILFIGTSTKALVKGIDTWKKETIMKKEAFEEAAQMLESGSTPDYASEEDYKSLPAD 213
+LLIILFI TSTKA KGIDTWKKETI KK EA+++LE+ + S +DYKSLP+
Sbjct: 182 VLLIILFIATSTKATYKGIDTWKKETIAKK----EASKLLEA----EPKSGDDYKSLPSG 337
Query: 214 LQD---EEVPLLDNIHWKELSVLMYVWVAFLIVQILKTYSKTCSIEYWLLNSLQV 265
+ EE PLL NI+WKELS+L YVWVA IVQI K Y+K CSI++WLLN LQV
Sbjct: 338 PTESLFEEAPLLKNIYWKELSLLAYVWVAIFIVQIGKEYTKPCSIQFWLLNFLQV 502
>TC220160 similar to UP|Q6Z001 (Q6Z001) Membrane protein-like, partial (29%)
Length = 610
Score = 233 bits (593), Expect = 2e-61
Identities = 107/137 (78%), Positives = 125/137 (91%)
Frame = +3
Query: 26 SFFWYSGESSYERVWPEMKFGWRIVVGSIVGFFGAALGSVGGVGGGGIFIPMLTLIIGFD 85
+F W GES Y+ VWP+M+FGW+I++G+IVGFFGAA GSVGGVGGGGIF+PML+LIIGFD
Sbjct: 6 NFLWQPGESGYQHVWPDMEFGWQIILGTIVGFFGAAFGSVGGVGGGGIFVPMLSLIIGFD 185
Query: 86 PKSSTALSKCMITGAAGSTVYYNLRLRHPTLDMPLIDYDLALLFQPMLMLGISIGVAFNV 145
KSSTA+SKCMI GAA STVYYNL LRHPTLD+P+IDYDLALL QPMLMLGISIGVAFNV
Sbjct: 186 AKSSTAISKCMIMGAALSTVYYNLNLRHPTLDLPIIDYDLALLIQPMLMLGISIGVAFNV 365
Query: 146 MFADWMVTILLIILFIG 162
+ ADWMVT+LL++LF+G
Sbjct: 366 VVADWMVTMLLLVLFLG 416
>TC205486 similar to GB|AAO63445.1|28951043|BT005381 At2g25737 {Arabidopsis
thaliana;} , partial (31%)
Length = 842
Score = 221 bits (564), Expect = 4e-58
Identities = 108/161 (67%), Positives = 135/161 (83%)
Frame = +1
Query: 282 GTRVIASRGKEITWKFHKICLYCFCGIIAGMVSGLLGLGGGFILGPLFLELGIPPQVASA 341
GTR +GK+ T ++ +YC G++AG+V G+LGLGGGF++GPLFLELG+PPQV+SA
Sbjct: 16 GTR--REQGKDFT--VLQLIIYCVFGVLAGVVGGMLGLGGGFVMGPLFLELGVPPQVSSA 183
Query: 342 TSTFAMVFSSSMSVVQYYHLDRFPIPYASYLVLVATIAALTGQHVVRKIIAIFGRASIIV 401
T+TFAM FSSSMSV++YY L RFPIPYA Y VLVATIAA GQH+VRK+I +FGRAS+I+
Sbjct: 184 TATFAMTFSSSMSVIEYYLLKRFPIPYALYFVLVATIAAFVGQHIVRKLIILFGRASLII 363
Query: 402 FILAFTIFVSAISLGGVGIGNMVEKMENEEYMGFDNLCHHG 442
FILA TIFVSA+SLGGVGI NMV K++N EYMGF++LC +G
Sbjct: 364 FILASTIFVSAVSLGGVGIVNMVHKIDNHEYMGFEDLCKYG 486
>TC205270 similar to UP|Q6Z001 (Q6Z001) Membrane protein-like, partial (35%)
Length = 1224
Score = 178 bits (452), Expect(2) = 1e-57
Identities = 91/185 (49%), Positives = 118/185 (63%), Gaps = 34/185 (18%)
Frame = +1
Query: 221 LLDNIHWKELSVLMYVWVAFLIVQILK--------------------------------- 247
+L+N+ WK L VL VWV L +I K
Sbjct: 40 ILENVRWKALGVLFTVWVLILASEIAKVGRYEISNSNPEFPFCFSSYVE**YNYAFVFKQ 219
Query: 248 TYSKTCSIEYWLLNSLQVPIAISVTLFEAICLCKGTRVIASRGKEIT-WKFHKICLYCFC 306
+++ TCS+EYW+LN LQVP+A+ T ++A+ L G RVI S+ + T W+ H++ +YC C
Sbjct: 220 SHTTTCSVEYWVLNLLQVPVALGATSYQAVLLYTGKRVITSKRDQRTQWRAHQLIMYCSC 399
Query: 307 GIIAGMVSGLLGLGGGFILGPLFLELGIPPQVASATSTFAMVFSSSMSVVQYYHLDRFPI 366
GI AG+V GLLGLGGG I+GPLFLELGIPPQV+SAT+TFAM FS+SMSVV+YY L RFPI
Sbjct: 400 GICAGIVGGLLGLGGGSIMGPLFLELGIPPQVSSATATFAMTFSASMSVVEYYLLKRFPI 579
Query: 367 PYASY 371
PY+ Y
Sbjct: 580 PYSKY 594
Score = 63.2 bits (152), Expect(2) = 1e-57
Identities = 32/48 (66%), Positives = 39/48 (80%)
Frame = +2
Query: 369 ASYLVLVATIAALTGQHVVRKIIAIFGRASIIVFILAFTIFVSAISLG 416
A Y V V+T AA GQ +VRK++AI GRAS+I+FIL+ TIFVSAISLG
Sbjct: 683 ALYFVAVSTFAAFVGQVLVRKLVAILGRASLIIFILSSTIFVSAISLG 826
Score = 44.7 bits (104), Expect = 9e-05
Identities = 18/34 (52%), Positives = 27/34 (78%)
Frame = +1
Query: 406 FTIFVSAISLGGVGIGNMVEKMENEEYMGFDNLC 439
F +F++ GGVGI NM++++ N+EYMGF+NLC
Sbjct: 919 FRVFIA----GGVGISNMIQRIANKEYMGFENLC 1008
>TC217984 similar to UP|Q8S9J0 (Q8S9J0) At2g36630/F1O11.26, partial (43%)
Length = 998
Score = 189 bits (480), Expect = 2e-48
Identities = 99/203 (48%), Positives = 132/203 (64%), Gaps = 7/203 (3%)
Frame = +3
Query: 244 QILKTYSKTCSIEYWLLNSLQVPIAISVTLFEAICLCKGTRVIASRGKE-------ITWK 296
Q++K + CS YW+L LQ+PIA+ V +EA+ L K + + G I W
Sbjct: 3 QVIKNDVEACSAWYWVLFGLQLPIALLVFGYEAVKLYKEHKRRMNTGNSECICEASIEWT 182
Query: 297 FHKICLYCFCGIIAGMVSGLLGLGGGFILGPLFLELGIPPQVASATSTFAMVFSSSMSVV 356
+ CGI+ G+V GLLG GGGF+L PL LE+G+ PQVASAT+TF M+FSSS+SVV
Sbjct: 183 AINLAFCALCGIVGGIVGGLLGSGGGFVLRPLLLEIGVIPQVASATATFVMMFSSSLSVV 362
Query: 357 QYYHLDRFPIPYASYLVLVATIAALTGQHVVRKIIAIFGRASIIVFILAFTIFVSAISLG 416
++Y L RFPIPYA YL V+ +A GQ VR++I GRASIIVFIL+ IF SA+++G
Sbjct: 363 EFYLLKRFPIPYALYLTSVSVLAGFWGQFFVRRLITCLGRASIIVFILSGVIFASALTMG 542
Query: 417 GVGIGNMVEKMENEEYMGFDNLC 439
VGI N ++ + N E+MGF C
Sbjct: 543 VVGIENSIQMINNHEFMGFLGFC 611
Score = 28.5 bits (62), Expect = 6.4
Identities = 25/80 (31%), Positives = 38/80 (47%), Gaps = 2/80 (2%)
Frame = +3
Query: 33 ESSYERVWPEMKFGWRIVVGSIVGFFGAALGSVGGVGGGGIFIPMLTLIIGFDPK--SST 90
E+S E W + + + G + G G LGS GGG + P+L L IG P+ S+T
Sbjct: 162 EASIE--WTAINLAFCALCGIVGGIVGGLLGS----GGGFVLRPLL-LEIGVIPQVASAT 320
Query: 91 ALSKCMITGAAGSTVYYNLR 110
A M + + +Y L+
Sbjct: 321 ATFVMMFSSSLSVVEFYLLK 380
>BI321835
Length = 433
Score = 128 bits (321), Expect(2) = 4e-47
Identities = 58/93 (62%), Positives = 72/93 (77%)
Frame = +3
Query: 22 MKKTSFFWYSGESSYERVWPEMKFGWRIVVGSIVGFFGAALGSVGGVGGGGIFIPMLTLI 81
+K F W Y+ VWPEM+F WRIVVG+++G GAA GSVGGVGGGGIF+PML LI
Sbjct: 15 IKALEFIWK--HLGYQHVWPEMEFSWRIVVGTLIGILGAAFGSVGGVGGGGIFVPMLILI 188
Query: 82 IGFDPKSSTALSKCMITGAAGSTVYYNLRLRHP 114
IGFDPKS+ A+SKCM+TGAA S V++ ++ RHP
Sbjct: 189 IGFDPKSAVAISKCMVTGAAISAVFFCMKQRHP 287
Score = 78.6 bits (192), Expect(2) = 4e-47
Identities = 39/49 (79%), Positives = 42/49 (85%)
Frame = +1
Query: 114 PTLDMPLIDYDLALLFQPMLMLGISIGVAFNVMFADWMVTILLIILFIG 162
PTLD P+IDYDL LL QP LMLGISIGV +V+FADWMVTILLIIL IG
Sbjct: 286 PTLDEPVIDYDLMLLIQPTLMLGISIGVILSVIFADWMVTILLIILCIG 432
>BE190433
Length = 472
Score = 164 bits (414), Expect = 1e-40
Identities = 84/155 (54%), Positives = 113/155 (72%), Gaps = 1/155 (0%)
Frame = +2
Query: 286 IASRGKEIT-WKFHKICLYCFCGIIAGMVSGLLGLGGGFILGPLFLELGIPPQVASATST 344
IAS+G + T W+ H + +YC CGI AG+V GLLGLGGG I PL L L IP + SAT+T
Sbjct: 2 IASKGDQRTQWRAHHLIMYCSCGICAGIVGGLLGLGGGHI*RPLLLALRIPAEECSATAT 181
Query: 345 FAMVFSSSMSVVQYYHLDRFPIPYASYLVLVATIAALTGQHVVRKIIAIFGRASIIVFIL 404
FAM FS+SM +V+YY L R PI Y YL V TIAA GQ VVRK++AI RAS+I++IL
Sbjct: 182 FAMTFSASMYLVEYYLLKRCPIAYTLYLPAVYTIAAFVGQVVVRKLVAILRRASLIIYIL 361
Query: 405 AFTIFVSAISLGGVGIGNMVEKMENEEYMGFDNLC 439
+++I V+AI LGGV I +M++++ +++ M +NLC
Sbjct: 362 SYSILVNAILLGGVDISHMIQELADDKTMSSENLC 466
>AI441331
Length = 330
Score = 159 bits (403), Expect = 2e-39
Identities = 76/106 (71%), Positives = 90/106 (84%)
Frame = +1
Query: 299 KICLYCFCGIIAGMVSGLLGLGGGFILGPLFLELGIPPQVASATSTFAMVFSSSMSVVQY 358
++ L+C CG +AGM++GLLGLGGGFILGPLFL LGIPPQVASATST M FS+SM+VV+Y
Sbjct: 10 QLVLFCACGTLAGMIAGLLGLGGGFILGPLFLGLGIPPQVASATSTLVMAFSASMAVVEY 189
Query: 359 YHLDRFPIPYASYLVLVATIAALTGQHVVRKIIAIFGRASIIVFIL 404
Y L RFP+PYA Y V +AT AAL GQH+VRK IAI GRAS+I+FIL
Sbjct: 190 YLLKRFPVPYALYFVAIATAAALVGQHLVRKAIAILGRASVIIFIL 327
>BI424298
Length = 422
Score = 150 bits (380), Expect = 9e-37
Identities = 73/139 (52%), Positives = 100/139 (71%), Gaps = 1/139 (0%)
Frame = +3
Query: 274 FEAICLCKGTRVIASRGKE-ITWKFHKICLYCFCGIIAGMVSGLLGLGGGFILGPLFLEL 332
+E L KG R ++S G T ++ +YC G++ +V GLL +GGGF++GPLFL L
Sbjct: 6 YEVPSLYKGRRQVSSVGDPGKTPTSQELTIYCIFGVLDAIVGGLLAIGGGFVMGPLFL*L 185
Query: 333 GIPPQVASATSTFAMVFSSSMSVVQYYHLDRFPIPYASYLVLVATIAALTGQHVVRKIIA 392
G+PPQV+SAT FAM FSSSM VV+YY L RFP+PYA Y ++VA I+ + GQ++VR +I
Sbjct: 186 GVPPQVSSATDNFAMTFSSSMCVVEYYLLKRFPVPYALYFIVVAAISEIVGQYIVRNLID 365
Query: 393 IFGRASIIVFILAFTIFVS 411
+ GR S+I+F+LAFTI VS
Sbjct: 366 VLGRVSLIIFVLAFTILVS 422
>TC205487 similar to GB|AAO63445.1|28951043|BT005381 At2g25737 {Arabidopsis
thaliana;} , partial (22%)
Length = 1048
Score = 150 bits (378), Expect = 1e-36
Identities = 71/102 (69%), Positives = 88/102 (85%)
Frame = +1
Query: 338 VASATSTFAMVFSSSMSVVQYYHLDRFPIPYASYLVLVATIAALTGQHVVRKIIAIFGRA 397
V+SAT+TFAM FSSSMSVV+YY L RFP+PYA Y + VA I+A+ GQH+VRK+I + GRA
Sbjct: 334 VSSATATFAMTFSSSMSVVEYYLLKRFPVPYALYFIAVAAISAIVGQHIVRKLIDVLGRA 513
Query: 398 SIIVFILAFTIFVSAISLGGVGIGNMVEKMENEEYMGFDNLC 439
S+I+F+LAFTI VSA+SLGGVGI MV+K+EN EYMGFD+LC
Sbjct: 514 SLIIFVLAFTILVSAVSLGGVGIVAMVKKIENHEYMGFDDLC 639
>AW099768
Length = 380
Score = 138 bits (347), Expect(2) = 2e-35
Identities = 63/78 (80%), Positives = 73/78 (92%)
Frame = +3
Query: 106 YYNLRLRHPTLDMPLIDYDLALLFQPMLMLGISIGVAFNVMFADWMVTILLIILFIGTST 165
YYNL LRHPTLD+P+IDYDLALL QPMLMLGISIGVAFNV+ ADWMVT+LL++LF+GTST
Sbjct: 3 YYNLNLRHPTLDLPIIDYDLALLIQPMLMLGISIGVAFNVVVADWMVTMLLLVLFLGTST 182
Query: 166 KALVKGIDTWKKETIMKK 183
KA KG++TWKKETIMK+
Sbjct: 183 KAFFKGVETWKKETIMKE 236
Score = 29.6 bits (65), Expect(2) = 2e-35
Identities = 14/41 (34%), Positives = 23/41 (55%), Gaps = 8/41 (19%)
Frame = +1
Query: 197 STPDYASEEDYKSLPA--------DLQDEEVPLLDNIHWKE 229
+T SE +Y +P+ D + EEV +L+N++WKE
Sbjct: 256 ATNGVGSEVEYTPIPSGPGSDIAKDTRHEEVSMLENVYWKE 378
>CO984940
Length = 549
Score = 126 bits (317), Expect = 2e-29
Identities = 67/142 (47%), Positives = 89/142 (62%), Gaps = 37/142 (26%)
Frame = -1
Query: 337 QVASATSTFAMVFSSSMSVVQYYHLDRFPIPY---------------------------- 368
QVASAT+TF M++SSS+SV+QYY L+RFP+PY
Sbjct: 543 QVASATATFGMMYSSSLSVIQYYLLNRFPVPYGKIKLNSHCTLLLFLNGFQRQHKLIQVI 364
Query: 369 ---------ASYLVLVATIAALTGQHVVRKIIAIFGRASIIVFILAFTIFVSAISLGGVG 419
A +L LVA IAA GQ+++ K++ IF RAS+I+F+LAFTIFVS+I+LGGVG
Sbjct: 363 FMHVSTCFAALFLTLVAAIAAFLGQYLIDKLVNIFQRASLIIFVLAFTIFVSSIALGGVG 184
Query: 420 IGNMVEKMENEEYMGFDNLCHH 441
I NM+ K++ EYMGFDN C +
Sbjct: 183 ISNMILKIQRNEYMGFDNFCRN 118
>TC223522 weakly similar to UP|Q6Z001 (Q6Z001) Membrane protein-like, partial
(18%)
Length = 429
Score = 122 bits (307), Expect = 2e-28
Identities = 55/90 (61%), Positives = 75/90 (83%)
Frame = +2
Query: 352 SMSVVQYYHLDRFPIPYASYLVLVATIAALTGQHVVRKIIAIFGRASIIVFILAFTIFVS 411
++SV+QYY L+RFP+PYA +L LVA IAA GQ+++ K++ IF RAS+I+F+LAFTIFVS
Sbjct: 8 ALSVIQYYLLNRFPVPYALFLTLVAAIAAFLGQYLIDKLVNIFQRASLIIFVLAFTIFVS 187
Query: 412 AISLGGVGIGNMVEKMENEEYMGFDNLCHH 441
+I+LGGVGI NM+ K++ EYMGFDN C +
Sbjct: 188 SIALGGVGISNMILKIQRNEYMGFDNFCRN 277
>AI795109
Length = 139
Score = 68.9 bits (167), Expect = 4e-12
Identities = 32/44 (72%), Positives = 37/44 (83%)
Frame = +2
Query: 100 AAGSTVYYNLRLRHPTLDMPLIDYDLALLFQPMLMLGISIGVAF 143
AA STV YNLR HPTLD+P+IDYD+ALL+QPML LG +IG AF
Sbjct: 5 AAVSTVCYNLRFTHPTLDLPVIDYDMALLYQPMLTLGTNIGAAF 136
>BF008683
Length = 142
Score = 66.2 bits (160), Expect = 3e-11
Identities = 31/46 (67%), Positives = 39/46 (84%)
Frame = +2
Query: 328 LFLELGIPPQVASATSTFAMVFSSSMSVVQYYHLDRFPIPYASYLV 373
LF LG+PPQV+SA+++FAM FSSSMSVV+YY L RFP+PYA Y +
Sbjct: 2 LF*VLGVPPQVSSASASFAMSFSSSMSVVEYYLLLRFPVPYALYFI 139
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.326 0.142 0.431
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,958,329
Number of Sequences: 63676
Number of extensions: 303398
Number of successful extensions: 2537
Number of sequences better than 10.0: 64
Number of HSP's better than 10.0 without gapping: 2474
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2520
length of query: 447
length of database: 12,639,632
effective HSP length: 100
effective length of query: 347
effective length of database: 6,272,032
effective search space: 2176395104
effective search space used: 2176395104
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC147964.5


