

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147964.4 + phase: 0 /pseudo
(276 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
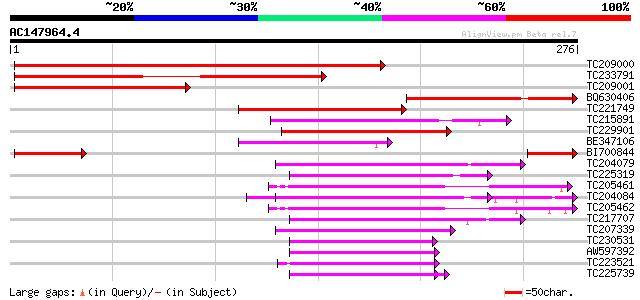
Score E
Sequences producing significant alignments: (bits) Value
TC209000 similar to UP|RU17_ARATH (Q42404) U1 small nuclear ribo... 341 2e-94
TC233791 similar to GB|AAD12776.1|1354466|ATU52910 U1 snRNP 70K ... 202 1e-52
TC209001 similar to GB|AAD12776.1|1354466|ATU52910 U1 snRNP 70K ... 166 8e-42
BQ630406 153 9e-38
TC221749 weakly similar to UP|Q16560 (Q16560) U1-snRNP binding p... 82 3e-16
TC215891 similar to GB|AAM78058.1|21928059|AY125548 AT5g61030/ma... 65 3e-11
TC229901 weakly similar to UP|GRP1_SORBI (Q99069) Glycine-rich R... 64 7e-11
BE347106 63 2e-10
BI700844 similar to SP|Q42404|RU17_ U1 small nuclear ribonucleop... 61 5e-10
TC204079 similar to UP|Q9SWA8 (Q9SWA8) Glycine-rich RNA-binding ... 59 2e-09
TC225319 similar to UP|P93486 (P93486) Glycine-rich RNA-binding ... 57 7e-09
TC205461 similar to GB|AAN28790.1|23505861|AY143851 At5g64200/MS... 56 2e-08
TC204084 UP|Q9SWA8 (Q9SWA8) Glycine-rich RNA-binding protein, co... 56 2e-08
TC205462 similar to GB|AAN28790.1|23505861|AY143851 At5g64200/MS... 56 2e-08
TC217707 similar to UP|Q941H8 (Q941H8) RNA-binding protein precu... 56 2e-08
TC207339 similar to UP|Q84ZB9 (Q84ZB9) RNA recognition motif (RR... 55 3e-08
TC230531 weakly similar to UP|Q941H9 (Q941H9) RNA-binding protei... 55 3e-08
AW597392 weakly similar to GP|7630030|emb| RNA binding protein-l... 55 5e-08
TC223521 weakly similar to UP|Q84ZB9 (Q84ZB9) RNA recognition mo... 54 6e-08
TC225739 homologue to UP|Q41124 (Q41124) Chloroplast RNA binding... 54 8e-08
>TC209000 similar to UP|RU17_ARATH (Q42404) U1 small nuclear
ribonucleoprotein 70 kDa (U1 snRNP 70 kDa) (snRNP70)
(U1-70K), partial (42%)
Length = 689
Score = 341 bits (874), Expect = 2e-94
Identities = 163/181 (90%), Positives = 173/181 (95%)
Frame = +3
Query: 3 DPLLRNQNAAVQARTKAQNRANVLQLKLIGQSHPTGLTANLLKLFEPRPPLEYKPPPEKR 62
D +RNQNAAVQARTKAQNR+NVLQLKLIGQSHPTGLTANLLKLFEPRPPLEYKPPPEKR
Sbjct: 147 DAFMRNQNAAVQARTKAQNRSNVLQLKLIGQSHPTGLTANLLKLFEPRPPLEYKPPPEKR 326
Query: 63 KCPPLTGMAQFVSKFAEPGEPEYSPPVPVVETPAERRARVHKLRLEKGAAKAAEELEKYD 122
KCPPL+GMAQFVSKFAEPGEPEY+PPVP ETPA++RAR+HKLRLEKGAAKAAEELEKYD
Sbjct: 327 KCPPLSGMAQFVSKFAEPGEPEYAPPVPETETPAQKRARIHKLRLEKGAAKAAEELEKYD 506
Query: 123 PHNDPNISGDPYKTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIE 182
PHNDPN+SGDPYKTLFVA+LSYETTESRIKREFESYG IKRVRLV D + NKPRGYAFIE
Sbjct: 507 PHNDPNVSGDPYKTLFVAKLSYETTESRIKREFESYGPIKRVRLVADKDINKPRGYAFIE 686
Query: 183 Y 183
Y
Sbjct: 687 Y 689
>TC233791 similar to GB|AAD12776.1|1354466|ATU52910 U1 snRNP 70K truncated
protein {Arabidopsis thaliana;} , partial (58%)
Length = 423
Score = 202 bits (514), Expect = 1e-52
Identities = 104/152 (68%), Positives = 115/152 (75%)
Frame = +3
Query: 3 DPLLRNQNAAVQARTKAQNRANVLQLKLIGQSHPTGLTANLLKLFEPRPPLEYKPPPEKR 62
DPL+RNQNAAVQAR K QNRAN LQLKLIGQSHPTGLTANLL+LFEPRP L+YKPPPEKR
Sbjct: 48 DPLIRNQNAAVQARAKDQNRANALQLKLIGQSHPTGLTANLLRLFEPRPALDYKPPPEKR 227
Query: 63 KCPPLTGMAQFVSKFAEPGEPEYSPPVPVVETPAERRARVHKLRLEKGAAKAAEELEKYD 122
A++RAR+HKLRLEKGAAKAAEELEKYD
Sbjct: 228 NAH---------------------------R**AQKRARIHKLRLEKGAAKAAEELEKYD 326
Query: 123 PHNDPNISGDPYKTLFVARLSYETTESRIKRE 154
PHNDPN+SGDPYKTLF+ARLSYE+T+SR+KRE
Sbjct: 327 PHNDPNVSGDPYKTLFIARLSYESTDSRMKRE 422
>TC209001 similar to GB|AAD12776.1|1354466|ATU52910 U1 snRNP 70K truncated
protein {Arabidopsis thaliana;} , partial (42%)
Length = 397
Score = 166 bits (421), Expect = 8e-42
Identities = 77/86 (89%), Positives = 83/86 (95%)
Frame = +1
Query: 3 DPLLRNQNAAVQARTKAQNRANVLQLKLIGQSHPTGLTANLLKLFEPRPPLEYKPPPEKR 62
D +RNQNAAVQARTKAQNR+NVLQLKLIGQSHPTGLTANLLKLFEPRPPLEYKPPPEKR
Sbjct: 139 DAFMRNQNAAVQARTKAQNRSNVLQLKLIGQSHPTGLTANLLKLFEPRPPLEYKPPPEKR 318
Query: 63 KCPPLTGMAQFVSKFAEPGEPEYSPP 88
KCPP +GMAQFVSKFA+PG+PEY+PP
Sbjct: 319 KCPPXSGMAQFVSKFAQPGDPEYAPP 396
>BQ630406
Length = 328
Score = 153 bits (386), Expect = 9e-38
Identities = 76/83 (91%), Positives = 79/83 (94%)
Frame = +1
Query: 194 KQADGRKIEGRRVLVDVERGRTVPNWRPRRLGGGLGTTRVGGDEVNQRHSGREQQQSGPS 253
KQADGRKI+GRRVLVDVERGRTVPNWRPRRLGGGLGTTRVGG+EVNQRHSGREQQQ S
Sbjct: 7 KQADGRKIDGRRVLVDVERGRTVPNWRPRRLGGGLGTTRVGGEEVNQRHSGREQQQ---S 177
Query: 254 RSEEPRVREERHADRDREKSRER 276
RSEEPRVRE+RHADRDRE SRER
Sbjct: 178 RSEEPRVREDRHADRDREISRER 246
>TC221749 weakly similar to UP|Q16560 (Q16560) U1-snRNP binding protein
homolog, isoform a (U11/U12 snRNP 35K), partial (34%)
Length = 422
Score = 82.0 bits (201), Expect = 3e-16
Identities = 39/82 (47%), Positives = 51/82 (61%)
Frame = +3
Query: 112 AKAAEELEKYDPHNDPNISGDPYKTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAE 171
A+ + YDP DP +GDPY TLFVARLS TTE +++ YG +K +RLV D
Sbjct: 123 AQLCSSIGLYDPFGDPKATGDPYCTLFVARLSRLTTEDDLRKVMSKYGRVKNLRLVRDIV 302
Query: 172 SNKPRGYAFIEYLHTRDMKAAY 193
+ RGYAF+EY R+M+ AY
Sbjct: 303 TGASRGYAFVEYETEREMRRAY 368
>TC215891 similar to GB|AAM78058.1|21928059|AY125548 AT5g61030/maf19_30
{Arabidopsis thaliana;} , partial (41%)
Length = 782
Score = 65.5 bits (158), Expect = 3e-11
Identities = 37/121 (30%), Positives = 57/121 (46%), Gaps = 4/121 (3%)
Frame = +2
Query: 128 NISGDPYKTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTR 187
++S P LF+ +SY T E ++ F YG + R++ D E+ + RG+ FI Y
Sbjct: 152 SMSSAPSTKLFIGGVSYSTDEQSLREAFSKYGEVVDARIIMDRETGRSRGFGFITYTSVE 331
Query: 188 DMKAAYKQADGRKIEGRRVLVDVERGRTVPNWRPRRLGGG----LGTTRVGGDEVNQRHS 243
+ +A + DG+ + GR + V+ N RPR GGG G GG E +
Sbjct: 332 EASSAIQALDGQDLHGRPIRVN------YANERPRGYGGGGFGSYGAVGGGGYEGGSSYR 493
Query: 244 G 244
G
Sbjct: 494 G 496
>TC229901 weakly similar to UP|GRP1_SORBI (Q99069) Glycine-rich RNA-binding
protein 1 (Fragment), partial (28%)
Length = 673
Score = 63.9 bits (154), Expect = 7e-11
Identities = 28/83 (33%), Positives = 52/83 (61%)
Frame = +3
Query: 133 PYKTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDMKAA 192
P L+V+ LS+ TTE ++ F+++G + V+LV D +N+PRG+AF+ Y + + A
Sbjct: 261 PQTKLYVSGLSFRTTEESLRNAFKNFGQLVEVKLVMDRIANRPRGFAFLRYATEEESQKA 440
Query: 193 YKQADGRKIEGRRVLVDVERGRT 215
+ G+ ++GR + V+V + R+
Sbjct: 441 IEGMHGKFLDGRVIFVEVAKPRS 509
>BE347106
Length = 472
Score = 62.8 bits (151), Expect = 2e-10
Identities = 33/76 (43%), Positives = 42/76 (54%), Gaps = 1/76 (1%)
Frame = +3
Query: 112 AKAAEELEKYDPHNDPNISGDPYKTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAE 171
A+ + YDP DP +GDPY TLFVARLS T E +++ YG +K RLV D
Sbjct: 111 AQLCSSIGLYDPFGDPRAAGDPYCTLFVARLSRLTNEDNLRKVMSKYGRVKNFRLVRDIG 290
Query: 172 SNKPRG-YAFIEYLHT 186
+K G Y F+ L T
Sbjct: 291 RSKNYGIYQFLRNLDT 338
>BI700844 similar to SP|Q42404|RU17_ U1 small nuclear ribonucleoprotein 70
kDa (U1 snRNP 70 kDa) (snRNP70) (U1-70K). [Mouse-ear
cress], partial (14%)
Length = 421
Score = 61.2 bits (147), Expect = 5e-10
Identities = 29/35 (82%), Positives = 32/35 (90%)
Frame = +2
Query: 3 DPLLRNQNAAVQARTKAQNRANVLQLKLIGQSHPT 37
D +RNQNAAVQA +KAQNR+NVLQLKLIGQSHPT
Sbjct: 152 DAFMRNQNAAVQASSKAQNRSNVLQLKLIGQSHPT 256
Score = 47.4 bits (111), Expect = 7e-06
Identities = 22/24 (91%), Positives = 23/24 (95%)
Frame = +3
Query: 253 SRSEEPRVREERHADRDREKSRER 276
SRSEEPRVRE+RHADRDRE SRER
Sbjct: 258 SRSEEPRVREDRHADRDREISRER 329
>TC204079 similar to UP|Q9SWA8 (Q9SWA8) Glycine-rich RNA-binding protein,
complete
Length = 748
Score = 58.9 bits (141), Expect = 2e-09
Identities = 35/122 (28%), Positives = 58/122 (46%)
Frame = +1
Query: 130 SGDPYKTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDM 189
S D FV L++ T + ++R F YG I +++ D E+ + RG+ F+ + + M
Sbjct: 73 SSDVEYRCFVGGLAWATDDQALERAFSQYGEIVETKIINDRETGRSRGFGFVTFASEQSM 252
Query: 190 KAAYKQADGRKIEGRRVLVDVERGRTVPNWRPRRLGGGLGTTRVGGDEVNQRHSGREQQQ 249
K A + +G+ ++GR + V+ + R GGG G R GG R G +
Sbjct: 253 KDAIEGMNGQNLDGRNITVNEAQSRGGGGGGGGG-GGGYGGGRGGGYGGGGRGGGGGYNR 429
Query: 250 SG 251
SG
Sbjct: 430 SG 435
Score = 32.7 bits (73), Expect = 0.18
Identities = 18/50 (36%), Positives = 19/50 (38%)
Frame = -3
Query: 49 PRPPLEYKPPPEKRKCPPLTGMAQFVSKFAEPGEPEYSPPVPVVETPAER 98
P PPL PPP R PP + P P Y PP P P R
Sbjct: 443 PPPPLRLYPPPPPRPPPP----------YPPPRPPPYPPPPPPPPPPPPR 324
>TC225319 similar to UP|P93486 (P93486) Glycine-rich RNA-binding protein
PsGRBP, partial (88%)
Length = 1167
Score = 57.4 bits (137), Expect = 7e-09
Identities = 29/99 (29%), Positives = 49/99 (49%)
Frame = +3
Query: 137 LFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDMKAAYKQA 196
LF+ LSY + +K F +G + +++TD +S + RG+ F+ + + +A
Sbjct: 372 LFIGGLSYGVDDQSLKDAFSGFGDVVDAKVITDRDSGRSRGFGFVNFSNDESASSALSAM 551
Query: 197 DGRKIEGRRVLVDVERGRTVPNWRPRRLGGGLGTTRVGG 235
DG+ ++GR + V R P+ GGG G R GG
Sbjct: 552 DGKDLDGRSIRVSYANDRPS---GPQSGGGGGGGYRSGG 659
>TC205461 similar to GB|AAN28790.1|23505861|AY143851 At5g64200/MSJ1_4
{Arabidopsis thaliana;} , partial (69%)
Length = 1148
Score = 56.2 bits (134), Expect = 2e-08
Identities = 40/150 (26%), Positives = 67/150 (44%), Gaps = 2/150 (1%)
Frame = +3
Query: 127 PNISGDPYKTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHT 186
P+IS D Y +L V +++ TT + F+ YG + + + D + + RG+AF+ Y +
Sbjct: 120 PDIS-DTY-SLLVLNITFRTTADDLFPLFDKYGKVVDIFIPKDRRTGESRGFAFVRYKYA 293
Query: 187 RDMKAAYKQADGRKIEGRRVLVDVERGRTVPNWRPRRLGGGLGTTRVGGDEVNQRHSGRE 246
+ + A ++ DGR ++GR + V + G + H GR
Sbjct: 294 DEAQKAVERLDGRMVDGREITVQFAK---------------------YGPNAERIHKGRI 410
Query: 247 QQQSGPSRSEEPRVREERHAD--RDREKSR 274
+ S SR R +RH D RDR+ R
Sbjct: 411 IETSPRSRRSRSRSPRKRHRDDYRDRDYRR 500
>TC204084 UP|Q9SWA8 (Q9SWA8) Glycine-rich RNA-binding protein, complete
Length = 1937
Score = 56.2 bits (134), Expect = 2e-08
Identities = 31/106 (29%), Positives = 52/106 (48%)
Frame = +2
Query: 130 SGDPYKTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDM 189
S D FV L++ T + ++R F YG I +++ D E+ + RG+ F+ + + M
Sbjct: 1226 SADVEYRCFVGGLAWATDDHALERAFSQYGEIVETKIINDRETGRSRGFGFVTFASEQSM 1405
Query: 190 KAAYKQADGRKIEGRRVLVDVERGRTVPNWRPRRLGGGLGTTRVGG 235
K A +G+ ++GR + V+ + R GGG G R GG
Sbjct: 1406 KDAIGAMNGQNLDGRNITVNEAQSRGGGGGGG---GGGGGYNRGGG 1534
Score = 55.1 bits (131), Expect = 3e-08
Identities = 46/169 (27%), Positives = 75/169 (44%), Gaps = 8/169 (4%)
Frame = -1
Query: 116 EELEKYDPHNDPNISGDPYKTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKP 175
+E + + + P+ S D LFV L + +R+ FES G ++ V ++ D + +
Sbjct: 932 QEEDTFGDGDGPSFSRD--LKLFVGNLPFSVDSARLAELFESAGNVEVVEVIYDKTTGRS 759
Query: 176 RGYAFIEYLHTRDMKAAYKQADGRKIEGRRVLVDVERGRTVPNWRPRRLGGGLGTTRVGG 235
RG+ F+ + +AA KQ +G +++GR + V+ PR GG +R GG
Sbjct: 758 RGFGFVTMSSVEEAEAAAKQFNGYELDGRSLRVNSGPPPARNESAPRFRGGSSFGSRGGG 579
Query: 236 --DEVNQRHSGR------EQQQSGPSRSEEPRVREERHADRDREKSRER 276
D N+ H G + R + +V E R DRE R R
Sbjct: 578 PSDSENRVHVGNLAWGVDDVALESLFREQGKKVLEAR-VIYDRESGRSR 435
>TC205462 similar to GB|AAN28790.1|23505861|AY143851 At5g64200/MSJ1_4
{Arabidopsis thaliana;} , partial (54%)
Length = 1403
Score = 55.8 bits (133), Expect = 2e-08
Identities = 43/155 (27%), Positives = 71/155 (45%), Gaps = 5/155 (3%)
Frame = +3
Query: 127 PNISGDPYKTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHT 186
P+IS D Y +L V +++ TT + F+ YG + + + D + + RG+AF+ Y +
Sbjct: 117 PDIS-DTY-SLLVLNITFRTTADDLFPLFDKYGKVVDIFIPKDRRTGESRGFAFVRYKYA 290
Query: 187 RDMKAAYKQADGRKIEGRRVLVDVERGRTVPNWRPRRLGGGLGTTRVGGDEVNQRHSGR- 245
+ + A ++ DGR ++GR + V + G + H GR
Sbjct: 291 DEAQKAVERLDGRMVDGREITVQFAK---------------------YGPNAERIHKGRI 407
Query: 246 --EQQQSGPSRSEEPRVR-EERHADRD-REKSRER 276
+S SRS PR R + + DRD R +SR R
Sbjct: 408 IETSPRSRRSRSRSPRKRHRDDYKDRDYRRRSRSR 512
>TC217707 similar to UP|Q941H8 (Q941H8) RNA-binding protein precursor,
partial (35%)
Length = 1008
Score = 55.8 bits (133), Expect = 2e-08
Identities = 35/119 (29%), Positives = 58/119 (48%), Gaps = 4/119 (3%)
Frame = +1
Query: 137 LFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDMKAAYKQA 196
LFV +SY T + ++ YG + V+++ D E+ + RG+ FI + + D A +
Sbjct: 169 LFVGGISYSTDDMSLRESPARYGEVIDVKVIMDRETGRSRGFGFITFATSEDASYAIQGM 348
Query: 197 DGRKIEGRRVLVDVERGRTVPNWRP----RRLGGGLGTTRVGGDEVNQRHSGREQQQSG 251
DG+ + GRR+ V+ R+ P + R GG G R GG+ +SG + G
Sbjct: 349 DGQDLHGRRIRVNYATERSRPGFGGDGGYRGSGGSDGYNR-GGNYGGGYNSGSDGYNRG 522
>TC207339 similar to UP|Q84ZB9 (Q84ZB9) RNA recognition motif
(RRM)-containing protein-like, partial (63%)
Length = 838
Score = 55.5 bits (132), Expect = 3e-08
Identities = 26/88 (29%), Positives = 44/88 (49%)
Frame = +3
Query: 130 SGDPYKTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDM 189
+ +P LFV+ LS T R++ EF +G + R+VTD S +G+ F++Y D
Sbjct: 186 TAEPNPNLFVSGLSKRTNTERLREEFAKFGEVVHARVVTDRVSGYSKGFGFVQYATIEDA 365
Query: 190 KAAYKQADGRKIEGRRVLVDVERGRTVP 217
+ DG+ ++G + + R R P
Sbjct: 366 AKGIEGMDGKFLDGWVIFAEYARPRPPP 449
>TC230531 weakly similar to UP|Q941H9 (Q941H9) RNA-binding protein precursor,
partial (29%)
Length = 851
Score = 55.1 bits (131), Expect = 3e-08
Identities = 26/72 (36%), Positives = 43/72 (59%)
Frame = +3
Query: 137 LFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDMKAAYKQA 196
LFV LSY+T E ++ F +G I V+++ D + K RGY F+ ++ AA K+
Sbjct: 420 LFVTGLSYDTNEPILRDAFGQHGEIIEVKVICDHVTGKSRGYGFVRFVSETTAAAARKEM 599
Query: 197 DGRKIEGRRVLV 208
+G+ ++GRR+ V
Sbjct: 600 NGQILDGRRIRV 635
>AW597392 weakly similar to GP|7630030|emb| RNA binding protein-like
{Arabidopsis thaliana}, partial (78%)
Length = 392
Score = 54.7 bits (130), Expect = 5e-08
Identities = 24/73 (32%), Positives = 44/73 (59%)
Frame = +2
Query: 137 LFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDMKAAYKQA 196
LFV RLS+ TT+ ++K+ F +G + + L D + +P+G+ F+ + + + A K
Sbjct: 83 LFVHRLSFYTTQEQLKKLFSPFGLVTQADLALDPITKRPKGFGFVSFKSEIEAEKACKAM 262
Query: 197 DGRKIEGRRVLVD 209
+GR + GR +LV+
Sbjct: 263 NGRIVNGRLILVE 301
>TC223521 weakly similar to UP|Q84ZB9 (Q84ZB9) RNA recognition motif
(RRM)-containing protein-like, partial (57%)
Length = 651
Score = 54.3 bits (129), Expect = 6e-08
Identities = 26/79 (32%), Positives = 45/79 (56%)
Frame = +1
Query: 131 GDPYKTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDMK 190
G YK LFV LS+ TTE+ + F +YG + ++VTD S++ +G+ F+ + + +
Sbjct: 70 GIAYK-LFVGGLSFYTTENALSEAFSNYGQVIEAKIVTDRVSDRSKGFGFVTFASQDEAE 246
Query: 191 AAYKQADGRKIEGRRVLVD 209
A + G+ + GR + VD
Sbjct: 247 NAIEDMKGKTLNGRVIFVD 303
>TC225739 homologue to UP|Q41124 (Q41124) Chloroplast RNA binding protein
precursor, partial (85%)
Length = 1436
Score = 53.9 bits (128), Expect = 8e-08
Identities = 25/78 (32%), Positives = 44/78 (56%)
Frame = +3
Query: 137 LFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDMKAAYKQA 196
LFV LS+ T + + F+ YG + R++ D E+ + RGY F+ Y +M+AA
Sbjct: 777 LFVGNLSWSVTNEILTQAFQEYGTVVGARVLYDGETGRSRGYGFVCYSTKAEMEAALAAL 956
Query: 197 DGRKIEGRRVLVDVERGR 214
+ ++EGR + V + +G+
Sbjct: 957 NDVELEGRAMRVSLAQGK 1010
Score = 41.6 bits (96), Expect = 4e-04
Identities = 18/73 (24%), Positives = 37/73 (50%)
Frame = +3
Query: 137 LFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDMKAAYKQA 196
L+ L Y +++ + YG+ + + ++ D ++ K RG+AF+ D A +
Sbjct: 504 LYFGNLPYSVDSAKLAGLIQDYGSAELIEVLYDRDTGKSRGFAFVTMSCIEDCNAVIENL 683
Query: 197 DGRKIEGRRVLVD 209
DG++ GR + V+
Sbjct: 684 DGKEFLGRTLRVN 722
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.313 0.133 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,592,797
Number of Sequences: 63676
Number of extensions: 168323
Number of successful extensions: 2099
Number of sequences better than 10.0: 265
Number of HSP's better than 10.0 without gapping: 1576
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1933
length of query: 276
length of database: 12,639,632
effective HSP length: 96
effective length of query: 180
effective length of database: 6,526,736
effective search space: 1174812480
effective search space used: 1174812480
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC147964.4


