

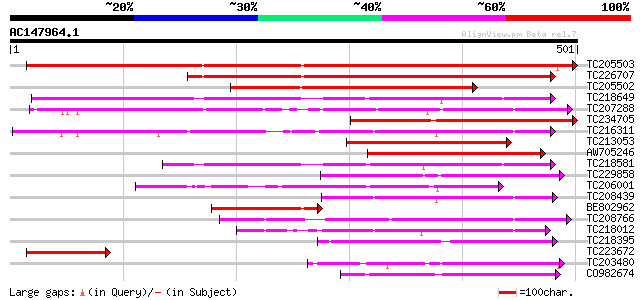
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147964.1 + phase: 0
(501 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC205503 UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, complete 580 e-166
TC226707 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter... 386 e-107
TC205502 homologue to UP|Q7XA50 (Q7XA50) Sorbitol-like transport... 258 4e-69
TC218649 similar to UP|Q39416 (Q39416) Integral membrane protein... 215 3e-56
TC207288 UP|Q7XA51 (Q7XA51) Monosaccharide transporter, complete 189 2e-48
TC234705 weakly similar to GB|CAD70577.1|32698459|MMU549317 solu... 176 2e-44
TC216311 UP|Q7XA52 (Q7XA52) Monosaccharide transporter, complete 173 2e-43
TC213053 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter... 169 3e-42
AW705246 158 5e-39
TC218581 similar to GB|AAM70554.1|21700861|AY124845 At1g75220/F2... 139 2e-33
TC229858 weakly similar to UP|Q8VHD6 (Q8VHD6) Glucose transporte... 135 4e-32
TC206001 similar to UP|Q8VZI5 (Q8VZI5) AT5g18840/F17K4_90, parti... 117 1e-26
TC208439 weakly similar to UP|O04078 (O04078) Monosaccharid tran... 114 1e-25
BE802962 similar to GP|29691859|gb| sorbitol transporter {Malus ... 114 1e-25
TC208766 similar to UP|STA_RICCO (Q10710) Sugar carrier protein ... 113 2e-25
TC218012 similar to UP|Q6VEF2 (Q6VEF2) Monosaccharide transporte... 113 2e-25
TC218395 similar to UP|Q9FIF2 (Q9FIF2) Sugar transporter-like pr... 112 5e-25
TC223672 homologue to UP|Q7XA50 (Q7XA50) Sorbitol-like transport... 108 7e-24
TC203480 similar to UP|Q8LPQ8 (Q8LPQ8) AT4g35300/F23E12_140, par... 105 4e-23
CO982674 98 8e-21
>TC205503 UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, complete
Length = 1928
Score = 580 bits (1494), Expect = e-166
Identities = 292/490 (59%), Positives = 384/490 (77%), Gaps = 4/490 (0%)
Frame = +1
Query: 16 NKYAFACAIVASMVSIVSGYDTGVMSGAMLFIKEDLGISDTQQEVLAGILNLCALVGSLT 75
NKYAFACA++ASM SI+ GYD GVMSGA ++IK DL +SD Q E+L GI+NL +L+GS
Sbjct: 247 NKYAFACAMLASMTSILLGYDIGVMSGAAIYIKRDLKVSDEQIEILLGIINLYSLIGSCL 426
Query: 76 AGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALMIAPVYSA 135
AGRTSD+IGRRYTI L +F++G+ LMG+ P+Y+ LM GR V G+G+G+ALMIAPVY+A
Sbjct: 427 AGRTSDWIGRRYTIGLGGAIFLVGSTLMGFYPHYSFLMCGRFVAGIGIGYALMIAPVYTA 606
Query: 136 EISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAALPSFVVAFC 195
E+S ASSRGFLTS PE+ I GILLGYISNY K L+LK+GWR+MLG+ A+PS V+
Sbjct: 607 EVSPASSRGFLTSFPEVFINGGILLGYISNYGFSK-LTLKVGWRMMLGVGAIPSVVLTEG 783
Query: 196 ILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDENCNDETVKLPQ 255
+L MPESPRWLVM+G+LG+A+KVL + S++ +EA+LRL +IK AAG+ E+CND+ V++ +
Sbjct: 784 VLAMPESPRWLVMRGRLGEARKVLNKTSDSKEEAQLRLAEIKQAAGIPESCNDDVVQVNK 963
Query: 256 KSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSKEK 315
+S+ GEGVWKEL L PTP++R ++IAA+GIHFF+ A+G++AV+LYSPRIF KAGIT+
Sbjct: 964 QSN-GEGVWKELFLYPTPAIRHIVIAALGIHFFQQASGVDAVVLYSPRIFEKAGITNDTH 1140
Query: 316 LLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLTVVDKSNGNV 375
LLAT+ VG K VF++ A F LD++GRR LL S GGM++ L L +SLTV+D S +
Sbjct: 1141KLLATVAVGFVKTVFILAATFTLDRVGRRPLLLSSVGGMVLSLLTLAISLTVIDHSERKL 1320
Query: 376 LWALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSMT 435
+WA+ SI AYVA F+IG GPITWVYSSEIFPL+LRAQGA+ GVAVNRT +AVVSMT
Sbjct: 1321MWAVGSSIAMVLAYVATFSIGAGPITWVYSSEIFPLRLRAQGAAAGVAVNRTTSAVVSMT 1500
Query: 436 FISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVF----TKKSSGKNV 491
F+S+ +AITIGG+FF++ GI+ + W+FFY LPET+GK LE+ME F +K ++ K V
Sbjct: 1501FLSLTRAITIGGAFFLYCGIATVGWIFFYTVLPETRGKTLEDMEGSFGTFRSKSNASKAV 1680
Query: 492 AIEMDPIQKV 501
E + +V
Sbjct: 1681ENENGQVAQV 1710
>TC226707 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, partial
(62%)
Length = 2338
Score = 386 bits (992), Expect = e-107
Identities = 191/325 (58%), Positives = 249/325 (75%)
Frame = +3
Query: 158 ILLGYISNYVLGKYLSLKLGWRLMLGIAALPSFVVAFCILTMPESPRWLVMQGQLGKAKK 217
IL+GYISNY K L+L+LGWRLMLG+ A+PS ++ +L MPESPRWLV +G+LG+AK+
Sbjct: 3 ILIGYISNYGFSK-LALRLGWRLMLGVGAIPSILIGVAVLAMPESPRWLVAKGRLGEAKR 179
Query: 218 VLMQVSNTTQEAELRLKDIKIAAGLDENCNDETVKLPQKSHQGEGVWKELILRPTPSVRW 277
VL ++S + +EA LRL DIK AG+ ++C+D+ V + +++H G GVW+EL L PTP+VR
Sbjct: 180 VLYKISESEEEARLRLADIKDTAGIPQDCDDDVVLVSKQTH-GHGVWRELFLHPTPAVRH 356
Query: 278 MLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSKEKLLLATIGVGLTKIVFLVIALFL 337
+ IA++GIHFF ATGI+AV+LYSPRIF KAGI S LLAT+ VG K V +++A F
Sbjct: 357 IFIASLGIHFFAQATGIDAVVLYSPRIFEKAGIKSDNYRLLATVAVGFVKTVSILVATFF 536
Query: 338 LDKLGRRRLLQISTGGMIIGLTLLGLSLTVVDKSNGNVLWALILSIVATYAYVAFFNIGL 397
LD+ GRR LL S G+I+ L LGLSLTVVD S + WA+ LSI A +YVA F+IG
Sbjct: 537 LDRAGRRVLLLCSVSGLILSLLTLGLSLTVVDHSQTTLNWAVGLSIAAVLSYVATFSIGS 716
Query: 398 GPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSMTFISIYKAITIGGSFFMFAGISV 457
GPITWVYSSEIFPL+LRAQG +IG AVNR + V++MTF+S+ KAITIGG+FF+FAG++
Sbjct: 717 GPITWVYSSEIFPLRLRAQGVAIGAAVNRVTSGVIAMTFLSLQKAITIGGAFFLFAGVAA 896
Query: 458 LAWLFFYFFLPETKGKALEEMEMVF 482
+AW+F Y LPET+GK LEE+E F
Sbjct: 897 VAWIFHYTLLPETRGKTLEEIEKSF 971
>TC205502 homologue to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, partial
(42%)
Length = 659
Score = 258 bits (659), Expect = 4e-69
Identities = 126/218 (57%), Positives = 168/218 (76%)
Frame = +1
Query: 196 ILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDENCNDETVKLPQ 255
+L MPESPRWLVM+G+LG+A+KVL + S++ +EA+LRL +IK AAG+ E+CND+ V++ +
Sbjct: 7 VLAMPESPRWLVMRGRLGEARKVLNKTSDSREEAQLRLAEIKQAAGIPESCNDDVVQVTK 186
Query: 256 KSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSKEK 315
+S GEGVWKEL L PTP +R ++IAA+GIHFF+ A+G++AV+LYSPRIF KAGI
Sbjct: 187 RS-TGEGVWKELFLYPTPPIRHIVIAALGIHFFQQASGVDAVVLYSPRIFEKAGIKDDTH 363
Query: 316 LLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLTVVDKSNGNV 375
LLAT+ VG K VF++ A F LD++GRR LL S GGM++ L L +SLT++ S +
Sbjct: 364 KLLATVAVGFVKTVFILAATFTLDRVGRRPLLLSSVGGMVLSLLTLAISLTIIGHSERKL 543
Query: 376 LWALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKL 413
+WA+ LSI AYVA F+IG GPITWVYSSEIFPL+L
Sbjct: 544 MWAVALSIAMVLAYVATFSIGAGPITWVYSSEIFPLRL 657
>TC218649 similar to UP|Q39416 (Q39416) Integral membrane protein, partial
(93%)
Length = 1695
Score = 215 bits (548), Expect = 3e-56
Identities = 138/468 (29%), Positives = 242/468 (51%), Gaps = 5/468 (1%)
Frame = +2
Query: 20 FACAIVASMVSIVSGYDTGVMSGAMLFIKEDLGISDTQQEVLAGILNLCALVGSLTAGRT 79
FAC ++ ++ I G+ G S I DLG+S ++ + + N+ A+VG++ +G+
Sbjct: 176 FACVLIVALGPIQFGFTAGYTSPTQSAIINDLGLSVSEFSLFGSLSNVGAMVGAIASGQI 355
Query: 80 SDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRCVCGVGVGFALMIAPVYSAEISS 139
++YIGR+ ++ +ASI I+G + + + + L +GR + G GVG PVY AEIS
Sbjct: 356 AEYIGRKGSLMIASIPNIIGWLAISFAKDSSFLYMGRLLEGFGVGIISYTVPVYIAEISP 535
Query: 140 ASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAALPSFVVAFCILTM 199
+ RG L S+ +L + IGI+L Y+ L + + WR++ I LP ++ + +
Sbjct: 536 PNLRGGLVSVNQLSVTIGIMLAYL--------LGIFVEWRILAIIGILPCTILIPALFFI 691
Query: 200 PESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIA-AGLDENCNDETVKLPQKSH 258
PESPRWL G + + L + + + + +IK A A + L Q+ +
Sbjct: 692 PESPRWLAKMGMTEEFETSLQVLRGFDTDISVEVNEIKRAVASTNTRITVRFADLKQRRY 871
Query: 259 QGEGVWKELILRPTPSVRWM-LIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSKEKLL 317
W+ L+ +G+ + +GI V+ YS IFR AGI+S +
Sbjct: 872 ------------------WLPLMIGIGLLILQQLSGINGVLFYSSTIFRNAGISSSD--- 988
Query: 318 LATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLTV-VDKSNGNVL 376
AT GVG +++ + L+L DK GRR LL +S GM L ++ ++ + S + L
Sbjct: 989 AATFGVGAVQVLATSLTLWLADKSGRRLLLIVSATGMSFSLLVVAITFYIKASISETSSL 1168
Query: 377 WALI--LSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSM 434
+ ++ LS+V A V F++G+G + W+ SEI P+ ++ S+ N + +V++
Sbjct: 1169YGILSTLSLVGVVAMVIAFSLGMGAMPWIIMSEILPINIKGLAGSVATLANWLFSWLVTL 1348
Query: 435 TFISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVF 482
T ++ + GG+F ++A + L +F ++PETKGK +EE++ F
Sbjct: 1349T-ANMLLDWSSGGTFTIYAVVCALTVVFVTIWVPETKGKTIEEIQWSF 1489
>TC207288 UP|Q7XA51 (Q7XA51) Monosaccharide transporter, complete
Length = 1813
Score = 189 bits (481), Expect = 2e-48
Identities = 147/503 (29%), Positives = 242/503 (47%), Gaps = 23/503 (4%)
Frame = +1
Query: 18 YAFACAIVASMVSIVSGYDTGVMSGAML---FIKE---------DLGISDTQQ------- 58
+AF C +V ++ + GYD GV G F+KE + + +T
Sbjct: 103 FAFTC-VVGALGGSLFGYDLGVSGGVPSMDDFLKEFFPKVYRRKQMHLHETDYCKYDDQV 279
Query: 59 -EVLAGILNLCALVGSLTAGRTSDYIGRRYTIFLASILFILGAGLMGYGPNYAILMVGRC 117
+ L ALV + A + GR+ +I + ++ F+ GA L N A+L++GR
Sbjct: 280 LTLFTSSLYFSALVMTFFASFLTRKKGRKASIIVGALSFLAGAILNAAAKNIAMLIIGRV 459
Query: 118 VCGVGVGFALMIAPVYSAEISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLG 177
+ G G+GF P+Y +E++ A +RG + L + GIL+ + NY K G
Sbjct: 460 LLGGGIGFGNQAVPLYLSEMAPAKNRGAVNQLFQFTTCAGILIANLVNYFTEKI--HPYG 633
Query: 178 WRLMLGIAALPSFVVAFCILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIK 237
WR+ LG+A LP+F + + E+P LV QG+L KAK+VL ++ T+ E +D+K
Sbjct: 634 WRISLGLAGLPAFAMLVGGICCAETPNSLVEQGRLDKAKQVLQRIRG-TENVEAEFEDLK 810
Query: 238 IAAGLDENCNDETVKLPQKSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAV 297
A+ + VK P ++ L+ R ++I A+GI F+ TG ++
Sbjct: 811 EASE-----EAQAVKSP---------FRTLLKRKYRP--QLIIGALGIPAFQQLTGNNSI 942
Query: 298 MLYSPRIFRKAGITSKEKLLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIG 357
+ Y+P IF+ G + L + I G +V VI++FL+DK GRR+ + MI
Sbjct: 943 LFYAPVIFQSLGFGANASLFSSFITNG-ALLVATVISMFLVDKYGRRKFFLEAGFEMICC 1119
Query: 358 LTLLGLSLTV---VDKSNGNVLWALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLR 414
+ + G L V K G + A + +V + +V + GP+ W+ SE+FPL++R
Sbjct: 1120MIITGAVLAVNFGHGKEIGKGVSAFL--VVVIFLFVLAYGRSWGPLGWLVPSELFPLEIR 1293
Query: 415 AQGASIGVAVNRTMNAVVSMTFISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKA 474
+ SI V VN A+V+ F+ + G F +FA + + F +F LPETK
Sbjct: 1294SSAQSIVVCVNMIFTALVAQLFLMSLCHLKF-GIFLLFASLIIFMSFFVFFLLPETKKVP 1470
Query: 475 LEEMEMVFTKKSSGKNVAIEMDP 497
+EE+ ++F + + DP
Sbjct: 1471IEEIYLLFENHWFWRRFVTDQDP 1539
>TC234705 weakly similar to GB|CAD70577.1|32698459|MMU549317 solute carrier
family 2 (facilitated glucose transporter), member 12
{Mus musculus;} , partial (5%)
Length = 992
Score = 176 bits (447), Expect = 2e-44
Identities = 86/200 (43%), Positives = 139/200 (69%)
Frame = +1
Query: 302 PRIFRKAGITSKEKLLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLL 361
P IF+ AGI KLL AT+ VG+ K +F+++A+ L+DKLGR+ LL IST GM + L +
Sbjct: 1 PEIFQAAGIEDNSKLLAATVAVGVAKTIFILVAIILIDKLGRKPLLMISTIGMTVCLFCM 180
Query: 362 GLSLTVVDKSNGNVLWALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIG 421
G +L ++ K + +A+ L+I+ VAFF++GLGP+ WV +SEIFPL++RAQ +++G
Sbjct: 181 GATLALLGKGS----FAIALAILFVCGNVAFFSVGLGPVCWVLTSEIFPLRVRAQASALG 348
Query: 422 VAVNRTMNAVVSMTFISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMV 481
NR + +V+M+F+S+ +AI++ G+FF+FA IS LA F +PETKGK+LE++EM+
Sbjct: 349 AVANRVCSGLVAMSFLSVSEAISVAGTFFVFAAISALAIAFVVTLVPETKGKSLEQIEMM 528
Query: 482 FTKKSSGKNVAIEMDPIQKV 501
F + + +E+ ++++
Sbjct: 529 FQNEYEIQGKEMELGDVEQL 588
>TC216311 UP|Q7XA52 (Q7XA52) Monosaccharide transporter, complete
Length = 1925
Score = 173 bits (438), Expect = 2e-43
Identities = 138/508 (27%), Positives = 232/508 (45%), Gaps = 28/508 (5%)
Frame = +1
Query: 3 INQGDKEDQTNTFNKYAFACAIVASMVSIVSGYDTGVMSGAM------------LFIKED 50
I+ G ++ + + IVA+M ++ GYD G+ G +F K++
Sbjct: 115 ISNGGGKEYPGSLTPFVTVTCIVAAMGGLIFGYDIGISGGVTSMDPFLLKFFPSVFRKKN 294
Query: 51 LGISDTQQ--------EVLAGILNLCALVGSLTAGRTSDYIGRRYTIFLASILFILGAGL 102
+ Q + L L AL+ SL A + GR+ ++ +LF++GA +
Sbjct: 295 SDKTVNQYCQYDSQTLTMFTSSLYLAALLSSLVASTVTRRFGRKLSMLFGGLLFLVGALI 474
Query: 103 MGYGPNYAILMVGRCVCGVGVGFALMIA---PVYSAEISSASSRGFLTSLPELCIGIGIL 159
G+ + +L+VGR + G G+GFA + P+ + I S G L + GI
Sbjct: 475 NGFAQHVWMLIVGRILLGFGIGFANQVCATLPI*NGFIQI*RSIGTLAFNCQSQFGIP-- 648
Query: 160 LGYISNYVLGKYLSLKLGWRLMLGIAAL-PSFVVAFCILTMPESPRWLVMQGQLGKAKKV 218
G +L + GW++ + A+ P+ ++ L +P++P ++ +G KAK
Sbjct: 649 RGQCVELLLWLKSMVAWGWKIEVWEGAMVPALIITVGSLVLPDTPNSMIERGDREKAKAQ 828
Query: 219 LMQVSNTTQEAELRLKDIKIAAGLDENCNDETVKLPQKSHQGEGVWKELILRPTPSVRWM 278
L ++ +DE ND V + S Q E W+ L+ R R
Sbjct: 829 LQRIRGIDN--------------VDEEFND-LVAASESSSQVEHPWRNLLQR---KYRPH 954
Query: 279 LIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSKEKLLLATIGVGLTKIVFLVIALFLL 338
L AV I FF+ TGI +M Y+P +F G L+ A I G+ +V ++++ +
Sbjct: 955 LTMAVLIPFFQQLTGINVIMFYAPVLFSSIGFKDDAALMSAVI-TGVVNVVATCVSIYGV 1131
Query: 339 DKLGRRRLLQISTGGMIIGLTLLGLSLTVVDKSNGNV----LWALILSIVATYAYVAFFN 394
DK GRR L M+I ++ ++ ++GN W I+ ++ YV+ F
Sbjct: 1132DKWGRRALFLEGGVQMLICQAVVAAAIGAKFGTDGNPGDLPKWYAIVVVLFICIYVSAFA 1311
Query: 395 IGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSMTFISIYKAITIGGSFFMFAG 454
GP+ W+ SEIFPL++R+ SI V+VN +++ F+++ + G F FA
Sbjct: 1312WSWGPLGWLVPSEIFPLEIRSAAQSINVSVNMLFTFLIAQVFLTMLCHMKF-GLFLFFAF 1488
Query: 455 ISVLAWLFFYFFLPETKGKALEEMEMVF 482
++ F YFFLPETKG +EEM V+
Sbjct: 1489FVLIMTFFVYFFLPETKGIPIEEMGQVW 1572
>TC213053 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, partial
(27%)
Length = 440
Score = 169 bits (428), Expect = 3e-42
Identities = 84/146 (57%), Positives = 109/146 (74%)
Frame = +1
Query: 298 MLYSPRIFRKAGITSKEKLLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIG 357
+LYSP IF+KAG+ S + LLAT+ VG K VF+++A FLLD++GRR LL S GGM+
Sbjct: 1 VLYSPEIFKKAGLESDGEQLLATVAVGFAKTVFILVATFLLDRVGRRPLLLTSVGGMVFS 180
Query: 358 LTLLGLSLTVVDKSNGNVLWALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQG 417
L LGLSLTV+D S + WA+ LSI +YV+ F++G GPITWVYSSEIFPL+LRAQG
Sbjct: 181 LLTLGLSLTVIDHSRAVLKWAIGLSIGMVLSYVSTFSVGAGPITWVYSSEIFPLRLRAQG 360
Query: 418 ASIGVAVNRTMNAVVSMTFISIYKAI 443
A++GV VNR + V+SMTF+S+ I
Sbjct: 361 AAMGVVVNRVTSGVISMTFLSLSNKI 438
>AW705246
Length = 484
Score = 158 bits (400), Expect = 5e-39
Identities = 79/157 (50%), Positives = 109/157 (69%)
Frame = +2
Query: 317 LLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLTVVDKSNGNVL 376
+L ++ VG K V + +A LD+ GRR LL + G+I+ L LGLSLTVVD S +
Sbjct: 2 VLTSVAVGFDKTVCIWVATIFLDRAGRRVLLLCNVSGLILSLMTLGLSLTVVDHSQTTLN 181
Query: 377 WALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSMTF 436
A+ L I + +YVA F+I GPITWVY SEIFPL+L A G +IG AVN+ + V++MTF
Sbjct: 182 CAVGLRIDSVLSYVATFSIWSGPITWVYISEIFPLRLWAHGVAIGAAVNKVTSGVIAMTF 361
Query: 437 ISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGK 473
+ + KAITIGG+FF+FAG++ +AW+F Y +PE +GK
Sbjct: 362 LYLQKAITIGGAFFLFAGVAAVAWIFHYTLIPEARGK 472
>TC218581 similar to GB|AAM70554.1|21700861|AY124845 At1g75220/F22H5_6
{Arabidopsis thaliana;} , partial (66%)
Length = 1340
Score = 139 bits (351), Expect = 2e-33
Identities = 102/351 (29%), Positives = 176/351 (50%), Gaps = 4/351 (1%)
Frame = +2
Query: 136 EISSASSRGFLTSLPELCIGIGILLGYISNYVLGKYLSLKLGWRLMLGIAALPSFVVAFC 195
EI+ G L S+ +L I IGI+L Y+ L L + WR++ + LP V+
Sbjct: 5 EIAPNHLIGGLGSVNQLSITIGIMLAYL--------LGLFVNWRILAILGILPCTVLIPG 160
Query: 196 ILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIA-AGLDENCNDETVKLP 254
+ +PESPRWL G + + + L + + + + +IK + A + L
Sbjct: 161 LFFIPESPRWLAKMGMIDEFETSLQVLRGFDTDISVEVHEIKRSVASTGKRAAIRFADLK 340
Query: 255 QKSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSKE 314
+K + W + L+ +G+ + +GI ++ YS IF AGI+S E
Sbjct: 341 RKRY-----W------------FPLMVGIGLLVLQQLSGINGILFYSTTIFANAGISSSE 469
Query: 315 KLLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLS--LTVVDKSN 372
AT+G+G +++ I+ +L+DK GRR LL IS+ M + L ++ ++ L V +
Sbjct: 470 A---ATVGLGAVQVIATGISTWLVDKSGRRLLLIISSSVMTVSLLIVSIAFYLEGVVSED 640
Query: 373 GNVLWAL-ILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAV 431
++ L I+SIV A V F++GLGPI W+ SEI P+ ++ SI N ++
Sbjct: 641 SHLFSILGIVSIVGLVAMVIGFSLGLGPIPWLIMSEILPVNIKGLAGSIATMGNWLISWG 820
Query: 432 VSMTFISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVF 482
++MT ++ + GG+F ++ ++ F ++PETKG+ LEE++ F
Sbjct: 821 ITMT-ANLLLNWSSGGTFTIYTVVAAFTIAFIAMWVPETKGRTLEEIQFSF 970
>TC229858 weakly similar to UP|Q8VHD6 (Q8VHD6) Glucose transporter 10 (Mus
musculus 13 days embryo heart cDNA, RIKEN full-length
enriched library, clone:D330001H08 product:solute
carrier family 2 (facilitated glucose transporter),
member 10, full insert sequence), partial (4%)
Length = 715
Score = 135 bits (340), Expect = 4e-32
Identities = 73/216 (33%), Positives = 121/216 (55%)
Frame = +3
Query: 275 VRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSKEKLLLATIGVGLTKIVFLVIA 334
+R + G+ F+ TGI VM YSP I + AG + E LL ++ V ++
Sbjct: 6 IRLAFLVGAGLLAFQQFTGINTVMYYSPTIVQMAGFHANELALLLSLIVAGMNAAGTILG 185
Query: 335 LFLLDKLGRRRLLQISTGGMIIGLTLLGLSLTVVDKSNGNVLWALILSIVATYAYVAFFN 394
++L+D GR++L S GG+I+ L +L + S N L+ L++V Y+ FF+
Sbjct: 186 IYLIDHAGRKKLALSSLGGVIVSLVILAFAF-YKQSSTSNELYGW-LAVVGLALYIGFFS 359
Query: 395 IGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSMTFISIYKAITIGGSFFMFAG 454
G+GP+ W SSEI+P + R + V N +VS TF+SI + I IG +F +
Sbjct: 360 PGMGPVPWTLSSEIYPEEYRGICGGMSATVCWVSNLIVSETFLSIAEGIGIGSTFLIIGV 539
Query: 455 ISVLAWLFFYFFLPETKGKALEEMEMVFTKKSSGKN 490
I+V+A++F ++PETKG +E+E+++ +++ GKN
Sbjct: 540 IAVVAFVFVLVYVPETKGLTFDEVEVIWRERAWGKN 647
>TC206001 similar to UP|Q8VZI5 (Q8VZI5) AT5g18840/F17K4_90, partial (66%)
Length = 951
Score = 117 bits (293), Expect = 1e-26
Identities = 96/328 (29%), Positives = 153/328 (46%), Gaps = 3/328 (0%)
Frame = +3
Query: 112 LMVGRCVCGVGVGFALMIAPVYSAEISSASSRGFLTSLPELCIGIGILLGYISNYVLGKY 171
L +GR G G+G + PVY AEI+ + RG L + +L I G G +S ++LG
Sbjct: 39 LDMGRFFTGYGIGVISYVVPVYIAEIAPKNLRGGLATTNQLLIVTG---GSVS-FLLGSV 206
Query: 172 LSLKLGWRLMLGIAALPSFVVAFCILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAEL 231
++ WR + +P + + +PESPRWL G+ +E +L
Sbjct: 207 IN----WRELALAGLVPCICLLVGLCFIPESPRWLAKVGR--------------EKEFQL 332
Query: 232 RLKDIKIAAGLDENCNDETVKLPQKSHQGEGVWKELILRPTPSVR-WMLIAAVGIHFFEH 290
L ++ G + +DE ++ E + K +L S ++ VG+ +
Sbjct: 333 ALSRLR---GKHADISDEAAEILDYIETLESLPKTKLLDLFQSKYVHSVVIGVGLMACQQ 503
Query: 291 ATGIEAVMLYSPRIFRKAGITSKEKLLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQIS 350
+ GI + Y+ IF AG++S + TI +I F ++ L+DK GRR L+ +S
Sbjct: 504 SVGINGIGFYTAEIFVAAGLSSGKA---GTIAYACIQIPFTLLGAILMDKSGRRPLVMVS 674
Query: 351 TGGMIIGLTLLGLSLTVVDKSNGNVL--WALILSIVATYAYVAFFNIGLGPITWVYSSEI 408
G +G + + + D+S +L W IL+ Y+A F+IGLG + WV SEI
Sbjct: 675 AAGTFLGCFVAAFAFFLKDQS---LLPEWVPILAFAGVLIYIAAFSIGLGSVPWVIMSEI 845
Query: 409 FPLKLRAQGASIGVAVNRTMNAVVSMTF 436
FP+ L+ S+ V V VVS TF
Sbjct: 846 FPIHLKGTAGSLVVLVAWLGAWVVSYTF 929
>TC208439 weakly similar to UP|O04078 (O04078) Monosaccharid transport
protein, partial (43%)
Length = 936
Score = 114 bits (284), Expect = 1e-25
Identities = 74/213 (34%), Positives = 108/213 (49%), Gaps = 4/213 (1%)
Frame = +1
Query: 276 RWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSKEKLLLATIGVGLTKIVFLVIAL 335
R L+ A+ I FF+ TG+ + Y+P +FR G S L+ A I +G K V ++++
Sbjct: 1 RPQLVFAICIPFFQQFTGLNVITFYAPILFRTIGFGSGASLMSAVI-IGSFKPVSTLVSI 177
Query: 336 FLLDKLGRRRLLQISTGGMIIGLTLLGLSLTVVDKSNGNV----LWALILSIVATYAYVA 391
L+DK GRR L M+I ++ +++ V +NGN W I+ + YV+
Sbjct: 178 LLVDKFGRRTLFLEGGAQMLICQIIMTIAIAVTFGTNGNPGTLPKWYAIVVVGIICVYVS 357
Query: 392 FFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSMTFISIYKAITIGGSFFM 451
F GP+ W+ SEIFPL++R SI V VN ++ F S+ + G F
Sbjct: 358 GFAWSWGPLGWLIPSEIFPLEIRPAAQSITVGVNMISTFFIAQFFTSMLCHMKF-GLFIF 534
Query: 452 FAGISVLAWLFFYFFLPETKGKALEEMEMVFTK 484
F V+ F Y LPETKG LEEM MV+ K
Sbjct: 535 FGCFVVIMTTFIYKLLPETKGIPLEEMSMVWQK 633
>BE802962 similar to GP|29691859|gb| sorbitol transporter {Malus x
domestica}, partial (19%)
Length = 295
Score = 114 bits (284), Expect = 1e-25
Identities = 54/98 (55%), Positives = 77/98 (78%)
Frame = +2
Query: 179 RLMLGIAALPSFVVAFCILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKI 238
R+MLG+ A+PS ++ +L MPESPRWLVM+G+LG+A+ VL + S + +EA+LRL +IK
Sbjct: 2 RMMLGVGAIPSVLLTVGVLAMPESPRWLVMRGRLGEARNVLNKSSYSKEEAQLRLAEIKQ 181
Query: 239 AAGLDENCNDETVKLPQKSHQGEGVWKELILRPTPSVR 276
AAG+ E+CND+ V++ S+ GEGVWKEL L PTP++R
Sbjct: 182 AAGIPESCNDDVVQVYNHSY-GEGVWKELFLYPTPAIR 292
>TC208766 similar to UP|STA_RICCO (Q10710) Sugar carrier protein A, partial
(55%)
Length = 1202
Score = 113 bits (283), Expect = 2e-25
Identities = 83/313 (26%), Positives = 152/313 (48%), Gaps = 2/313 (0%)
Frame = +1
Query: 186 ALPSFVVAFCILTMPESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDEN 245
A+P+ ++ + +P++P L+ +G K +K+L ++ T +E + +D+ A+ L ++
Sbjct: 1 AVPALLMTVGGIFLPDTPNSLIERGLAEKGRKLLEKIRGT-KEVDAEFQDMVDASELAKS 177
Query: 246 CNDETVKLPQKSHQGEGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIF 305
+ ++ ++ E L+ A+ + F+ TGI +++ Y+P +F
Sbjct: 178 IKHPFRNILERRYRPE-----------------LVMAIFMPTFQILTGINSILFYAPVLF 306
Query: 306 RKAGITSKEKLLLATIGVG-LTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLGLS 364
+ G L+ + + G L F+ IA +D+LGRR LL MI ++ +
Sbjct: 307 QSMGFGGDASLISSALTGGVLASSTFISIAT--VDRLGRRVLLVSGGLQMITCQIIVAII 480
Query: 365 LTVVDKSNGNVLWAL-ILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVA 423
L V ++ + IL +V +V F GP+ W SEIFPL++R+ G I VA
Sbjct: 481 LGVKFGADQELSKGFSILVVVVICLFVVAFGWSWGPLGWTVPSEIFPLEIRSAGQGITVA 660
Query: 424 VNRTMNAVVSMTFISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVFT 483
VN +++ F+++ + G F FAG + +F Y FLPETKG +EEM ++
Sbjct: 661 VNLLFTFIIAQAFLALLCSFKF-GIFLFFAGWITIMTIFVYLFLPETKGIPIEEMSFMWR 837
Query: 484 KKSSGKNVAIEMD 496
+ K + + D
Sbjct: 838 RHWFWKRICLPTD 876
>TC218012 similar to UP|Q6VEF2 (Q6VEF2) Monosaccharide transporter 4, partial
(54%)
Length = 1142
Score = 113 bits (282), Expect = 2e-25
Identities = 82/280 (29%), Positives = 138/280 (49%), Gaps = 2/280 (0%)
Frame = +3
Query: 201 ESPRWLVMQGQLGKAKKVLMQVSNTTQEAELRLKDIKIAAGLDENCNDETVKLPQKSHQG 260
++P L+ +G+L + K VL ++ T EL +++ A+ + + VK P
Sbjct: 3 DTPNSLIERGRLEEGKTVLKKIRGT-DNIELEFQELVEASRVAKE-----VKHP------ 146
Query: 261 EGVWKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSKEKLLLAT 320
++ L+ R R L+ ++ + F+ TGI A+M Y+P +F G + L A
Sbjct: 147 ---FRNLLKRRN---RPQLVISIALQIFQQFTGINAIMFYAPVLFNTLGFKNDASLYSAV 308
Query: 321 IGVGLTKIVFLVIALFLLDKLGRRRLLQISTGGMIIGLTLLG--LSLTVVDKSNGNVLWA 378
I G ++ V++++ +DKLGRR LL + M + ++ L + V D S+
Sbjct: 309 I-TGAVNVLSTVVSIYSVDKLGRRMLLLEAGVQMFLSQVVIAIILGIKVTDHSDDLSKGI 485
Query: 379 LILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSMTFIS 438
IL +V +V+ F GP+ W+ SE FPL+ R+ G S+ V VN V++ F+S
Sbjct: 486 AILVVVMVCTFVSSFAWSWGPLGWLIPSETFPLETRSAGQSVTVCVNLLFTFVIAQAFLS 665
Query: 439 IYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEM 478
+ G F F+G ++ +F F LPETK +EEM
Sbjct: 666 MLCHFKF-GIFLFFSGWVLVMSVFVLFLLPETKNVPIEEM 782
>TC218395 similar to UP|Q9FIF2 (Q9FIF2) Sugar transporter-like protein,
partial (40%)
Length = 1013
Score = 112 bits (279), Expect = 5e-25
Identities = 68/212 (32%), Positives = 110/212 (51%)
Frame = +1
Query: 273 PSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSKEKLLLATIGVGLTKIVFLV 332
P+++ +I G+ F+ TG +V+ Y+ I + AG ++ ++ +GL K++
Sbjct: 109 PNLKAFIIGG-GLVLFQQITGQPSVLYYAGPILQSAGFSAASDATKVSVVIGLFKLLMTW 285
Query: 333 IALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLTVVDKSNGNVLWALILSIVATYAYVAF 392
IA+ +D LGRR LL G+ + L LL + + AL+L YV
Sbjct: 286 IAVLKVDDLGRRPLLIGGVSGIALSLVLLSAYYKFLGGFPLVAVGALLL-------YVGC 444
Query: 393 FNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNAVVSMTFISIYKAITIGGSFFMF 452
+ I GPI+W+ SE+FPL+ R +G S+ V N NAVV+ F + + + F +F
Sbjct: 445 YQISFGPISWLMVSEVFPLRTRGKGISLAVLTNFASNAVVTFAFSPLKEFLGAENLFLLF 624
Query: 453 AGISVLAWLFFYFFLPETKGKALEEMEMVFTK 484
I+ L+ LF F +PETKG +LE++E K
Sbjct: 625 GAIATLSLLFIIFSVPETKGMSLEDIESKILK 720
>TC223672 homologue to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, partial
(19%)
Length = 468
Score = 108 bits (269), Expect = 7e-24
Identities = 52/74 (70%), Positives = 62/74 (83%)
Frame = +1
Query: 16 NKYAFACAIVASMVSIVSGYDTGVMSGAMLFIKEDLGISDTQQEVLAGILNLCALVGSLT 75
NKYAFACA++ASM SI+ GYD GVMSGA L+IK DL +SD Q E+L GI+NL +L+GS
Sbjct: 238 NKYAFACAVLASMTSILLGYDIGVMSGAALYIKRDLKVSDVQIEILLGIINLYSLIGSCL 417
Query: 76 AGRTSDYIGRRYTI 89
AGRTSD+IGRRYTI
Sbjct: 418 AGRTSDWIGRRYTI 459
>TC203480 similar to UP|Q8LPQ8 (Q8LPQ8) AT4g35300/F23E12_140, partial (38%)
Length = 1040
Score = 105 bits (263), Expect = 4e-23
Identities = 74/244 (30%), Positives = 125/244 (50%), Gaps = 17/244 (6%)
Frame = +2
Query: 264 WKELILRPTPSVRWMLIAAVGIHFFEHATGIEAVMLYSPRIFRKAGITSKEKLLLATIGV 323
WK L+ P V+ L+ VGI + +GI V+ Y+P+I +AG+ ++LL+ IG+
Sbjct: 173 WKALL---EPGVKHALVVGVGIQILQQFSGINGVLYYTPQILEEAGV----EVLLSDIGI 331
Query: 324 GLTKIVFLV-------------IALFLLDKLGRRRLLQISTGGMIIGLTLLGLSLTVVDK 370
G FL+ +A+ L+D GRR+LL + +I+ L +L + V
Sbjct: 332 GSESASFLISAFTTFLMLPCIGVAMKLMDVSGRRQLLLTTIPVLIVSLIILVIGSLV--- 502
Query: 371 SNGNVLWALILSIVATYAYVAFFNIGLGPITWVYSSEIFPLKLRAQGASIGVAVNRTMNA 430
+ GNV A I S V Y F +G GPI + SEIFP ++R +I V +
Sbjct: 503 NFGNVAHAAI-STVCVVVYFCCFVMGYGPIPNILCSEIFPTRVRGLCIAICALVFWIGDI 679
Query: 431 VVSMTFISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETKGKALEEMEMVFT----KKS 486
+++ + + ++ +GG F ++A + ++W+F + +PETKG LE + F+ + +
Sbjct: 680 IITYSLPVMLGSLGLGGVFAIYAVVCFISWIFVFLKVPETKGMPLEVISEFFSVGAKQAA 859
Query: 487 SGKN 490
S KN
Sbjct: 860 SAKN 871
>CO982674
Length = 703
Score = 98.2 bits (243), Expect = 8e-21
Identities = 64/195 (32%), Positives = 102/195 (51%), Gaps = 1/195 (0%)
Frame = -1
Query: 293 GIEAVMLYSPRIFRKAGITSKEKLLLATIGVGLTKIVFLVIALFLLDKLGRRRLLQISTG 352
GI V+ +S F+K G+ S LA++ VGLT + AL+L+D+ GR++LL S
Sbjct: 691 GINGVLYFSSLTFQKVGVESSA---LASLFVGLTNFAGALCALYLIDREGRQKLLIGSYL 521
Query: 353 GMIIGLTLLGLSLTV-VDKSNGNVLWALILSIVATYAYVAFFNIGLGPITWVYSSEIFPL 411
GM I + L+ + +D+ GN L SI+ T Y+ F IG GP+T + E+
Sbjct: 520 GMAISMFLVASGIIFPLDEQLGNNL-----SILGTIMYIFSFAIGAGPVTGIIIPELSST 356
Query: 412 KLRAQGASIGVAVNRTMNAVVSMTFISIYKAITIGGSFFMFAGISVLAWLFFYFFLPETK 471
+ R + + + N VV + F+ + + + F IS+LA F Y+F+ ETK
Sbjct: 355 RTRGKIMGFSFSTHWVCNFVVGLFFLELVDKFGVAPVYASFGAISLLAATFAYYFIVETK 176
Query: 472 GKALEEMEMVFTKKS 486
G++LEE+E K+
Sbjct: 175 GRSLEEIERSLNLKA 131
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.325 0.141 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,688,317
Number of Sequences: 63676
Number of extensions: 286056
Number of successful extensions: 2284
Number of sequences better than 10.0: 115
Number of HSP's better than 10.0 without gapping: 2187
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2216
length of query: 501
length of database: 12,639,632
effective HSP length: 101
effective length of query: 400
effective length of database: 6,208,356
effective search space: 2483342400
effective search space used: 2483342400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC147964.1


