

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147961.2 - phase: 0 /pseudo
(451 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
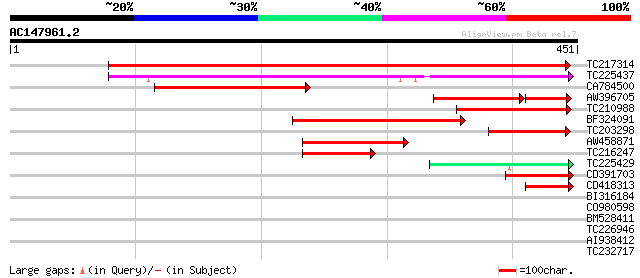
Score E
Sequences producing significant alignments: (bits) Value
TC217314 weakly similar to UP|O80938 (O80938) CER1-like protein,... 469 e-133
TC225437 similar to UP|Q8H1Z0 (Q8H1Z0) Cuticle protein (Faceless... 196 2e-50
CA784500 weakly similar to GP|1199467|dbj possible aldehyde deca... 167 9e-42
AW396705 similar to GP|1209703|gb|A maize gl1 homolog {Arabidops... 121 7e-40
TC210988 similar to UP|O80938 (O80938) CER1-like protein, partia... 156 2e-38
BF324091 weakly similar to GP|1418319|emb| CER1-like {Arabidopsi... 145 4e-35
TC203298 UP|CB23_SOYBN (P09756) Chlorophyll a-b binding protein ... 94 2e-19
AW458871 83 3e-16
TC216247 similar to UP|O04077 (O04077) Sucrose transport protein... 72 5e-13
TC225429 similar to UP|Q8H1Z0 (Q8H1Z0) Cuticle protein (Faceless... 53 3e-07
CD391703 50 2e-06
CD418313 43 3e-04
BI316184 similar to GP|4426627|gb| sterol-C5(6)-desaturase homol... 33 0.34
CO980598 32 0.77
BM528411 30 1.7
TC226946 similar to UP|IF39_TOBAC (P56821) Eukaryotic translatio... 30 2.9
AI938412 29 3.8
TC232717 similar to UP|Q8QHS4 (Q8QHS4) CMP3.5R (CMP204.5L), part... 29 3.8
>TC217314 weakly similar to UP|O80938 (O80938) CER1-like protein, partial
(50%)
Length = 1544
Score = 469 bits (1208), Expect = e-133
Identities = 216/368 (58%), Positives = 279/368 (75%)
Frame = +1
Query: 79 FHSLHHTKFRRNYSLFMPMYDYIYGTVDKSTDVIYETSLMRPKESPDVVHLTHLTTFNSI 138
FHSLHHT+FR NYSLFMP+YDYIYGT DK++D ++E++L + +E P+VVHLTHLTT SI
Sbjct: 184 FHSLHHTQFRTNYSLFMPLYDYIYGTTDKASDKLHESALKQEEEIPNVVHLTHLTTPESI 363
Query: 139 YQLRLGFASLASNPQTSKWYLHLMWPFTMFSMLMTWICGRAFVLESNSFKNLKLQCWLIP 198
Y LRLGFA LAS P TSKWYL LMWP T +SM++TW+ GR F++E N F LKLQ W IP
Sbjct: 364 YHLRLGFAYLASKPYTSKWYLCLMWPVTAWSMILTWVYGRTFIVEGNRFDKLKLQTWAIP 543
Query: 199 RFKRQYFSKWQSKTFNNLIEEAIVEAELNGAKVISLGLFNKNHQLNERHEHYIGRLPQLK 258
++ QYF + Q N +IEEAI++A+ G KV+SLGL N+ LN Y+ R P+LK
Sbjct: 544 KYSLQYFMQSQKVAINTMIEEAILDADRKGIKVLSLGLRNQGEDLNIYGGLYVSRHPKLK 723
Query: 259 IKVVDGSSLAAATVLNNIPKGTNQVLLRGKFNKVAFVIANALCKKNVQVVVLYKDELKEL 318
++VVDGSSL A VLN+IPKGT QVLLRGK K+A+ +A LC++ VQV LY+D+ L
Sbjct: 724 VRVVDGSSLVVAVVLNSIPKGTTQVLLRGKLTKIAYALAYTLCQQGVQVAALYEDDYVRL 903
Query: 319 EQRINTSKGNLALSPFNTPKIWLVGDEWDEYEQMEAPKGSLFIPFSHFPPKKMRKDCFYH 378
++ N+S+ NLA + +T WLVGD E EQ++APKG+LFIP++ FPP+K RKDCFYH
Sbjct: 904 KKSFNSSETNLAFTKSSTQTTWLVGDGLTEEEQLKAPKGTLFIPYTQFPPRKYRKDCFYH 1083
Query: 379 YTPAMITPTTFMNSHSCENWLPRRVMSAWRIAGIIHALEGWNVHECGDTILSTEKVWEAS 438
TPAM+ P + N HSCE+WLPRR+MSAWRIAGI+H+LEGW HECG T+ + + VW ++
Sbjct: 1084CTPAMLAPCSVENIHSCEDWLPRRIMSAWRIAGIVHSLEGWTEHECGHTMHNIDNVWHST 1263
Query: 439 IRHGFQPL 446
++HGFQPL
Sbjct: 1264LQHGFQPL 1287
>TC225437 similar to UP|Q8H1Z0 (Q8H1Z0) Cuticle protein (Faceless pollen-1)
(YORE-YORE protein), partial (70%)
Length = 1577
Score = 196 bits (498), Expect = 2e-50
Identities = 121/385 (31%), Positives = 194/385 (49%), Gaps = 15/385 (3%)
Frame = +3
Query: 79 FHSLHHTKFRRNYSLFMPMYDYIYGTVDKST---DVIYETSLMRPKESPDVVHLTHLTTF 135
+H LHH+ N+ LFMP++D + T++K++ + + P V L H+
Sbjct: 219 YHHLHHSDKDTNFCLFMPLFDSLGNTLNKNSWQSHKLLSSGSGNGDMVPHFVFLAHIVDV 398
Query: 136 NSIYQLRLGFASLASNPQTSKWYLHLMWPFTMFSMLMTWICGRAFVLESNSFKNLKLQCW 195
+S + + S AS P T++++L P T +L W + F++ + Q W
Sbjct: 399 SSSMHAQFVYRSFASLPYTTRFFLLPGLPITFLVLLAMWAWSKTFLVSFYYLRGRLHQTW 578
Query: 196 LIPRFKRQYFSKWQSKTFNNLIEEAIVEAELNGAKVISLGLFNKNHQLNERHEHYIGRLP 255
++PR QYF + ++ NN IE+AI+ A+ G KVISL NKN LN + ++ + P
Sbjct: 579 VVPRCGFQYFLPFATEGINNQIEQAILRADKIGVKVISLAALNKNESLNGGGKLFVDKHP 758
Query: 256 QLKIKVVDGSSLAAATVLNNIPKGTNQVLLRGKFNKVAFVIANALCKKNVQVVV--LYKD 313
L+++VV G++L AA +LN IP+ +V L G +K+ IA LC+K V+V++ L D
Sbjct: 759 NLRVRVVHGNTLTAAVILNEIPQDVKEVFLTGATSKLGRAIALYLCQKKVKVLMLTLSTD 938
Query: 314 ELKELEQR---------INTSKGNLALSPFNTPKIWLVGDEWDEYEQMEAPKGSLFIPFS 364
+ +++ + +K A K W+VG EQ AP+G+ F F
Sbjct: 939 RFQRIQKEAPPEYQSYLVQVTKYQAA----QNCKTWIVGKWITPREQYWAPRGTHFHQFV 1106
Query: 365 HFPPKKMRKDCFYHYTPAMITPTTFMNSHSCENWLPRRVMSAWRIAGIIHALEGWNVHEC 424
P RKDC Y AM P CE + R V+ A G++H+LEGW HE
Sbjct: 1107VPPILPFRKDCTYGDLAAMRLPEDVEGLGCCEYTMDRGVVHACHAGGVVHSLEGWPHHEV 1286
Query: 425 GD-TILSTEKVWEASIRHGFQPLKN 448
G + + VWEA+++HG +P+ +
Sbjct: 1287GAIDVNRIDLVWEAALKHGLRPVSS 1361
>CA784500 weakly similar to GP|1199467|dbj possible aldehyde decarbonylase
{Arabidopsis thaliana}, partial (18%)
Length = 396
Score = 167 bits (423), Expect = 9e-42
Identities = 80/124 (64%), Positives = 98/124 (78%)
Frame = +2
Query: 116 SLMRPKESPDVVHLTHLTTFNSIYQLRLGFASLASNPQTSKWYLHLMWPFTMFSMLMTWI 175
+L R + DVVHLTHLTT SIY LRLGFASLAS PQ+S WYL+LMWPFT++S+L+TW
Sbjct: 5 ALKREESLADVVHLTHLTTPESIYHLRLGFASLASRPQSSTWYLYLMWPFTLWSVLVTWF 184
Query: 176 CGRAFVLESNSFKNLKLQCWLIPRFKRQYFSKWQSKTFNNLIEEAIVEAELNGAKVISLG 235
G+ FV+E N+FK L LQ W+IPRF QY KWQS+T N LIEEAI++AEL+ KV+SLG
Sbjct: 185 YGQTFVMERNAFKMLNLQSWVIPRFHVQYLFKWQSETLNKLIEEAILQAELSKVKVLSLG 364
Query: 236 LFNK 239
L N+
Sbjct: 365 LSNQ 376
>AW396705 similar to GP|1209703|gb|A maize gl1 homolog {Arabidopsis
thaliana}, partial (11%)
Length = 422
Score = 121 bits (304), Expect(2) = 7e-40
Identities = 52/72 (72%), Positives = 61/72 (84%)
Frame = +1
Query: 338 KIWLVGDEWDEYEQMEAPKGSLFIPFSHFPPKKMRKDCFYHYTPAMITPTTFMNSHSCEN 397
KIWL+GD+ +E +Q +APKGSLFIP S FPPKK+RKDCFYH TPAMI P + +N SCEN
Sbjct: 34 KIWLLGDQCNEVDQRKAPKGSLFIPISQFPPKKLRKDCFYHSTPAMIAPPSLVNVDSCEN 213
Query: 398 WLPRRVMSAWRI 409
WLPRRVMSAWR+
Sbjct: 214 WLPRRVMSAWRV 249
Score = 60.8 bits (146), Expect(2) = 7e-40
Identities = 24/37 (64%), Positives = 33/37 (88%)
Frame = +3
Query: 411 GIIHALEGWNVHECGDTILSTEKVWEASIRHGFQPLK 447
GI+HALEGWNV+ECG+ + S EK+ +AS++HGF+PLK
Sbjct: 252 GILHALEGWNVNECGNEMFSVEKIRQASLQHGFRPLK 362
>TC210988 similar to UP|O80938 (O80938) CER1-like protein, partial (11%)
Length = 449
Score = 156 bits (394), Expect = 2e-38
Identities = 66/92 (71%), Positives = 79/92 (85%)
Frame = +1
Query: 356 KGSLFIPFSHFPPKKMRKDCFYHYTPAMITPTTFMNSHSCENWLPRRVMSAWRIAGIIHA 415
KGSLFIP S FPPKK+RKDCFYH TPAMI P + +N SCENWLPRRVMSAWR+AGI+HA
Sbjct: 10 KGSLFIPISQFPPKKLRKDCFYHSTPAMIAPPSLVNVDSCENWLPRRVMSAWRVAGILHA 189
Query: 416 LEGWNVHECGDTILSTEKVWEASIRHGFQPLK 447
LE W V+ECG+ + S EK+W+AS++HGF+PLK
Sbjct: 190 LECWKVNECGNVMFSVEKIWQASLQHGFRPLK 285
>BF324091 weakly similar to GP|1418319|emb| CER1-like {Arabidopsis thaliana},
partial (9%)
Length = 418
Score = 145 bits (366), Expect = 4e-35
Identities = 78/138 (56%), Positives = 97/138 (69%), Gaps = 1/138 (0%)
Frame = +3
Query: 226 LNGAKVISLGLFNKNHQLNERHEHYIGRLPQLKIKVVDGSSLAAATVLNNIPKGTNQVLL 285
L+ KV+SLGL N+ N+ E YI R P+LKIK+VDGSSL A V+N+IPK QVLL
Sbjct: 3 LSKVKVLSLGLSNQGDSFNKYGELYIKRYPELKIKIVDGSSLVVAIVVNSIPKEARQVLL 182
Query: 286 RGKFNKVAFVIANALCKKNVQVVVLYKDELKELEQRI-NTSKGNLALSPFNTPKIWLVGD 344
GK NKV++ IA+ALC++ +V +YKDE +L+ RI N SK NL T KIWLVGD
Sbjct: 183 CGKPNKVSYAIASALCERGTKVTTMYKDEYDKLQLRISNESKKNLVFPGSYTAKIWLVGD 362
Query: 345 EWDEYEQMEAPKGSLFIP 362
+ DE EQ +APKGSLFIP
Sbjct: 363 QCDEVEQKKAPKGSLFIP 416
>TC203298 UP|CB23_SOYBN (P09756) Chlorophyll a-b binding protein 3,
chloroplast precursor (LHCII type I CAB-3) (LHCP),
complete
Length = 1518
Score = 93.6 bits (231), Expect = 2e-19
Identities = 38/65 (58%), Positives = 52/65 (79%)
Frame = -1
Query: 382 AMITPTTFMNSHSCENWLPRRVMSAWRIAGIIHALEGWNVHECGDTILSTEKVWEASIRH 441
AM+ P+ N HSCE+WLPRRVMSAWRIAGI+H+LE W+ +EC + + +KVW ++++H
Sbjct: 456 AMLIPSCVENVHSCEDWLPRRVMSAWRIAGIVHSLERWSTNECNYKMHNIDKVWRSTLQH 277
Query: 442 GFQPL 446
GFQPL
Sbjct: 276 GFQPL 262
>AW458871
Length = 411
Score = 82.8 bits (203), Expect = 3e-16
Identities = 43/84 (51%), Positives = 57/84 (67%)
Frame = +1
Query: 234 LGLFNKNHQLNERHEHYIGRLPQLKIKVVDGSSLAAATVLNNIPKGTNQVLLRGKFNKVA 293
LGL N+ N+ E YI R P+LKIK+VDGSSL A V+N+IPK QVLL GK NKV+
Sbjct: 1 LGLSNQGDSFNKYGELYIKRYPELKIKIVDGSSLVVAIVVNSIPKEARQVLLCGKPNKVS 180
Query: 294 FVIANALCKKNVQVVVLYKDELKE 317
+ IA+ALC++ +V ++ E E
Sbjct: 181 YAIASALCERGTKVCLILFSEFVE 252
>TC216247 similar to UP|O04077 (O04077) Sucrose transport protein, partial
(17%)
Length = 930
Score = 72.0 bits (175), Expect = 5e-13
Identities = 37/58 (63%), Positives = 41/58 (69%)
Frame = +1
Query: 234 LGLFNKNHQLNERHEHYIGRLPQLKIKVVDGSSLAAATVLNNIPKGTNQVLLRGKFNK 291
LGL N+ LN Y+ R P LK+K+VDGSSLAAA VLNNIPKGT QVLL GK K
Sbjct: 757 LGLMNQGEDLNIYGGLYVSRNPNLKVKMVDGSSLAAAVVLNNIPKGTTQVLLMGKLTK 930
>TC225429 similar to UP|Q8H1Z0 (Q8H1Z0) Cuticle protein (Faceless pollen-1)
(YORE-YORE protein), partial (18%)
Length = 729
Score = 52.8 bits (125), Expect = 3e-07
Identities = 41/162 (25%), Positives = 59/162 (36%), Gaps = 48/162 (29%)
Frame = +1
Query: 335 NTPKIWLVGDEWDEYEQMEAPKGSLFIPFSHFPPKKMRKDCFYHYTPAMITPTTFMNSHS 394
N+ W+VG EQ AP+G+ F F P RKDC Y AM P
Sbjct: 1 NSAPAWIVGKWITPREQYWAPRGTHFHQFVVPPILSFRKDCTYGDLAAMRLPEDVEGLGC 180
Query: 395 CE-----------------------------------------------NWLPRRVMSAW 407
CE + R V+ A
Sbjct: 181 CEVKPSSTLAVSVSFIFLLIDMI*FFSHEPITYFYLLTLINICLIRYMQYTMDRGVVHAC 360
Query: 408 RIAGIIHALEGWNVHECGD-TILSTEKVWEASIRHGFQPLKN 448
G++H+LEGW+ HE G + + VWEA+++HG +P+ +
Sbjct: 361 HAGGVVHSLEGWSHHEVGAIDVNRIDLVWEAALKHGLRPVSS 486
>CD391703
Length = 358
Score = 50.1 bits (118), Expect = 2e-06
Identities = 21/55 (38%), Positives = 35/55 (63%), Gaps = 1/55 (1%)
Frame = -3
Query: 395 CENWLPRRVMSAWRIAGIIHALEGWNVHECGD-TILSTEKVWEASIRHGFQPLKN 448
CE + R V+ A G++H+LEGW+ HE G + + VWEA+++HG +P+ +
Sbjct: 254 CEYTMDRGVVHACHAGGVVHSLEGWSHHEVGAIDVNRIDLVWEAALKHGLRPVSS 90
>CD418313
Length = 670
Score = 42.7 bits (99), Expect = 3e-04
Identities = 16/39 (41%), Positives = 28/39 (71%), Gaps = 1/39 (2%)
Frame = -1
Query: 411 GIIHALEGWNVHECGD-TILSTEKVWEASIRHGFQPLKN 448
G++H+LEGW+ HE G + + VWEA+++HG +P+ +
Sbjct: 382 GVVHSLEGWSHHEVGAIDVNRIDLVWEAALKHGLRPVSS 266
>BI316184 similar to GP|4426627|gb| sterol-C5(6)-desaturase homolog
{Nicotiana tabacum}, partial (48%)
Length = 421
Score = 32.7 bits (73), Expect = 0.34
Identities = 10/27 (37%), Positives = 19/27 (70%)
Frame = +2
Query: 79 FHSLHHTKFRRNYSLFMPMYDYIYGTV 105
+H++HHT +R NY + D+++GT+
Sbjct: 281 YHTIHHTTYRHNYGHYTIWMDWMFGTL 361
>CO980598
Length = 733
Score = 31.6 bits (70), Expect = 0.77
Identities = 12/25 (48%), Positives = 16/25 (64%)
Frame = -3
Query: 80 HSLHHTKFRRNYSLFMPMYDYIYGT 104
HSLHH++F NY F +D + GT
Sbjct: 497 HSLHHSRFEVNYGQFFTAFDRLGGT 423
>BM528411
Length = 368
Score = 30.4 bits (67), Expect = 1.7
Identities = 11/26 (42%), Positives = 17/26 (65%)
Frame = +1
Query: 79 FHSLHHTKFRRNYSLFMPMYDYIYGT 104
+H LHH+ N+ LFMP++D + T
Sbjct: 280 YHHLHHSDKDTNFCLFMPLFDSLGNT 357
>TC226946 similar to UP|IF39_TOBAC (P56821) Eukaryotic translation initiation
factor 3 subunit 9 (eIF-3 eta) (eIF3 p110) (eIF3b),
partial (18%)
Length = 646
Score = 29.6 bits (65), Expect = 2.9
Identities = 25/64 (39%), Positives = 34/64 (53%), Gaps = 6/64 (9%)
Frame = -1
Query: 36 SNIACWTSRVFILLAS*STSSSLSILSLSF-----SPSFFNCH*ACHLFH-SLHHTKFRR 89
S++ TS + AS S+SSSL+ SLSF S S+ H HL H S + +F R
Sbjct: 382 SSVRSITSSTSMSFASYSSSSSLASPSLSFCLCKLSSSWIRFHSLTHLSHSSFNILRFSR 203
Query: 90 NYSL 93
+ SL
Sbjct: 202 SCSL 191
>AI938412
Length = 181
Score = 29.3 bits (64), Expect = 3.8
Identities = 13/36 (36%), Positives = 18/36 (49%)
Frame = +1
Query: 389 FMNSHSCENWLPRRVMSAWRIAGIIHALEGWNVHEC 424
+M S C W V+ W+ A IHA + WN+ C
Sbjct: 73 YMLSIPCRIWYL*TVLCRWKAAYTIHASDDWNL*LC 180
>TC232717 similar to UP|Q8QHS4 (Q8QHS4) CMP3.5R (CMP204.5L), partial (22%)
Length = 1102
Score = 29.3 bits (64), Expect = 3.8
Identities = 13/33 (39%), Positives = 19/33 (57%)
Frame = -2
Query: 69 FFNCH*ACHLFHSLHHTKFRRNYSLFMPMYDYI 101
F + H CH++HSL F N + ++P Y YI
Sbjct: 1089 FLSLHIXCHIYHSL----FLLNLNPYIPQYIYI 1003
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.328 0.139 0.455
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,784,072
Number of Sequences: 63676
Number of extensions: 413322
Number of successful extensions: 3134
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 3099
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3125
length of query: 451
length of database: 12,639,632
effective HSP length: 100
effective length of query: 351
effective length of database: 6,272,032
effective search space: 2201483232
effective search space used: 2201483232
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC147961.2


