

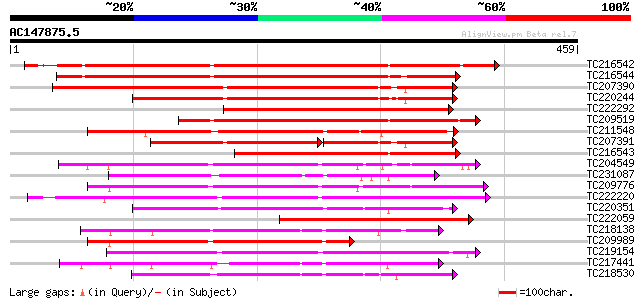
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147875.5 - phase: 0
(459 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC216542 weakly similar to UP|Q9LL55 (Q9LL55) Receptor-like kina... 357 6e-99
TC216544 weakly similar to UP|Q9LL55 (Q9LL55) Receptor-like kina... 337 7e-93
TC207390 weakly similar to UP|Q9ATQ5 (Q9ATQ5) LRK33, partial (34%) 306 1e-83
TC220244 weakly similar to UP|Q9LL58 (Q9LL58) Receptor-like kina... 274 5e-74
TC222292 similar to UP|Q9FID5 (Q9FID5) Receptor protein kinase-l... 256 1e-68
TC209519 similar to UP|Q84K89 (Q84K89) Receptor kinase LRK10 (Re... 241 6e-64
TC211548 weakly similar to UP|Q7XIP7 (Q7XIP7) Receptor-like kina... 239 1e-63
TC207391 similar to UP|Q9SWU8 (Q9SWU8) Receptor-like kinase (Fra... 156 4e-61
TC216543 weakly similar to UP|Q84V32 (Q84V32) Receptor kinase LR... 210 1e-54
TC204549 similar to UP|Q9M0X5 (Q9M0X5) Receptor protein kinase-l... 207 8e-54
TC231087 similar to UP|Q39203 (Q39203) Receptor-like protein kin... 199 3e-51
TC209776 similar to UP|O65470 (O65470) Serine/threonine kinase-l... 198 5e-51
TC222220 weakly similar to UP|Q9ZP16 (Q9ZP16) Receptor-like prot... 197 6e-51
TC220351 similar to UP|Q961E7 (Q961E7) GH28523p (CG1830-PB) , pa... 197 6e-51
TC222059 similar to GB|CAD39676.1|21741394|OSJN00121 OSJNBb0051N... 184 7e-47
TC218138 homologue to UP|Q8GRK2 (Q8GRK2) Somatic embryogenesis r... 181 6e-46
TC209989 similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kin... 180 1e-45
TC219154 similar to PIR|T05754|T05754 S-receptor kinase M4I22.1... 179 3e-45
TC217441 homologue to GB|AAK68074.1|14573459|AF384970 somatic em... 177 7e-45
TC218530 similar to UP|Q9SGU0 (Q9SGU0) T6H22.8.1 protein, partia... 174 1e-43
>TC216542 weakly similar to UP|Q9LL55 (Q9LL55) Receptor-like kinase, partial
(38%)
Length = 1435
Score = 357 bits (916), Expect = 6e-99
Identities = 179/384 (46%), Positives = 256/384 (66%)
Frame = +2
Query: 13 FGVTMFFIIFISCYFKKGIRRPQMTIFRKRRKHVDNNVEVFMQSYNLSIARRYSYAEVKR 72
FG+T+F ++ I +K +RKR + N+E +++ NL + YSY E+K+
Sbjct: 263 FGMTLFIVLLI---YK----------WRKRHLSIYENIENYLEQNNL-MPIGYSYKEIKK 400
Query: 73 ITNSFRDKLGHGGYGVVYKASLTDGRQVAVKVINESKGNGEEFINEVASISRTSHMNIVS 132
+ F++KLG GGYG+V+K L G VA+K+++++KGNG++FI+E+A+I R H N+V
Sbjct: 401 MARGFKEKLGGGGYGIVFKGKLHSGPSVAIKMLHKAKGNGQDFISEIATIGRIHHQNVVQ 580
Query: 133 LLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQ 192
L+GYC E +KRAL+YEFMP GSLDKFI+ D + ++ IAIG+ARG+ YLH
Sbjct: 581 LIGYCVEGSKRALVYEFMPNGSLDKFIFTK---DGNIHLTYDEIYNIAIGVARGIAYLHH 751
Query: 193 GCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRGTIGYMAPEVFS 252
GC +ILH DIKP NILLDE F PK+SDFGLAK+ +++SIV++ RGTIGYMAPE+F
Sbjct: 752 GCEMQILHFDIKPHNILLDETFTPKVSDFGLAKLYPIDNSIVTMTAARGTIGYMAPELFY 931
Query: 253 RAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYFPDWIYKDLEQGNDLLNSLT 312
+ GG+S+K+DVYS+GML++EM RKN +S++YFP WIY L + D +
Sbjct: 932 KNIGGISHKADVYSFGMLLMEMASKRKNLNPHADHSSQLYFPFWIYNQLGKETD-IEMEG 1108
Query: 313 ISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQGPLSSVTFPPKPVLFSPKRPPL 372
++EEEN + KK+ +VSLWCIQ P DRP MNKV+EML+G + S+ PPKP L+ P
Sbjct: 1109VTEEENKIAKKMIIVSLWCIQLKPTDRPSMNKVVEMLEGDIESLEIPPKPSLY-PHETME 1285
Query: 373 QLSNMSSSDWQETNSITTETELEE 396
++ SS T+ I++ +E
Sbjct: 1286NDQSIYSSQTMSTDFISSSNYSKE 1357
>TC216544 weakly similar to UP|Q9LL55 (Q9LL55) Receptor-like kinase, partial
(36%)
Length = 1229
Score = 337 bits (864), Expect = 7e-93
Identities = 166/327 (50%), Positives = 226/327 (68%)
Frame = +2
Query: 39 FRKRRKHVDNNVEVFMQSYNLSIARRYSYAEVKRITNSFRDKLGHGGYGVVYKASLTDGR 98
+RKR + ++E +++ NL + RYSY EVK++ F+DKLG GGYG V+K L G
Sbjct: 233 WRKRHLSMFESIENYLEQNNL-MPIRYSYKEVKKMAGGFKDKLGEGGYGSVFKGKLGSGS 409
Query: 99 QVAVKVINESKGNGEEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKF 158
VA+K++ +SKGNG++FI+EVA+I RT H NIV L+G+C +KRAL+YEFMP GSLDKF
Sbjct: 410 CVAIKMLGKSKGNGQDFISEVATIGRTYHQNIVQLIGFCVHGSKRALVYEFMPNGSLDKF 589
Query: 159 IYKSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKI 218
I+ D + ++ I+I +ARG+ YLH GC +ILH DIKP NILLDENF PK+
Sbjct: 590 IFSK---DESIHLSYDRIYNISIEVARGIAYLHYGCEMQILHFDIKPHNILLDENFTPKV 760
Query: 219 SDFGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGR 278
SDFGLAK+ +++SIV RGTIGYMAPE+F GG+S+K+DVYSYGML++EM R
Sbjct: 761 SDFGLAKLYPIDNSIVPRTAARGTIGYMAPELFYNNIGGISHKADVYSYGMLLMEMASKR 940
Query: 279 KNYQTGGSCTSEMYFPDWIYKDLEQGNDLLNSLTISEEENDMVKKITMVSLWCIQTNPLD 338
KN +S+++FP WIY + D + ++EEE KK+ +V+LWCIQ P D
Sbjct: 941 KNLNPHAERSSQLFFPFWIYNHIGDEED-IEMEDVTEEE----KKMIIVALWCIQLKPND 1105
Query: 339 RPPMNKVIEMLQGPLSSVTFPPKPVLF 365
RP MNKV+EML+G + ++ PPKP L+
Sbjct: 1106RPSMNKVVEMLEGDIENLEIPPKPTLY 1186
>TC207390 weakly similar to UP|Q9ATQ5 (Q9ATQ5) LRK33, partial (34%)
Length = 1342
Score = 306 bits (784), Expect = 1e-83
Identities = 152/331 (45%), Positives = 221/331 (65%), Gaps = 3/331 (0%)
Frame = +3
Query: 35 QMTIFRKRRKHVDNNVEVFMQSYNLSIARRYSYAEVKRITNSFRDKLGHGGYGVVYKASL 94
++ ++ ++++ V F++ Y R++YA++KRIT F++KLG G +G V++ L
Sbjct: 123 KIALYFRQKEEDQARVAKFLEDYRAEKPARFTYADLKRITGGFKEKLGEGAHGAVFRGKL 302
Query: 95 TDGRQVAVKVINESKGNGEEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGS 154
++ VAVK++N ++G G+EFINEV + + H+N+V LLG+C E RAL+Y P GS
Sbjct: 303 SNEILVAVKILNNTEGEGKEFINEVGIMGKIHHINVVRLLGFCAEGFHRALVYNLFPNGS 482
Query: 155 LDKFIYKSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENF 214
L +FI D + L QIA+GIA+G+EYLH+GC+ I+H DI P N+LLD+NF
Sbjct: 483 LQRFIVPPDDKDHFLGWEK--LQQIALGIAKGIEYLHEGCNQPIIHFDINPHNVLLDDNF 656
Query: 215 CPKISDFGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEM 274
PKISDFGLAK+C N S+VS+ RGT+GY+APEVFS+ FG VSYKSD+YSYGML+LEM
Sbjct: 657 TPKISDFGLAKLCSKNPSLVSMTAARGTLGYIAPEVFSKNFGNVSYKSDIYSYGMLLLEM 836
Query: 275 IGGRKNYQTGGSCTSEMYFPDWIYKDLEQGNDLLNSLTISEEEND--MVKKITMVSLWCI 332
+GGRKN + + +PDWI+ +L G+ ++ E+E D + KK+ +V LWCI
Sbjct: 837 VGGRKNVDMSSAQDFHVLYPDWIH-NLIDGDVHIH----VEDECDIKIAKKLAIVGLWCI 1001
Query: 333 QTNPLDRPPMNKVIEMLQ-GPLSSVTFPPKP 362
Q P++RP + VI+ML+ G S + PP P
Sbjct: 1002QWQPVNRPSIKSVIQMLETGGESQLNVPPNP 1094
>TC220244 weakly similar to UP|Q9LL58 (Q9LL58) Receptor-like kinase, partial
(32%)
Length = 1112
Score = 274 bits (701), Expect = 5e-74
Identities = 135/265 (50%), Positives = 187/265 (69%), Gaps = 2/265 (0%)
Frame = +1
Query: 100 VAVKVINESKGNGEEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFI 159
VAVK++N++ G+G++FINEV ++ + H+N+V LLG+C + RAL+Y+F P GSL +F+
Sbjct: 40 VAVKILNDTVGDGKDFINEVGTMGKIHHVNVVRLLGFCADGFHRALVYDFFPNGSLQRFL 219
Query: 160 YKSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKIS 219
D + L QIA+G+A+G+EYLH GC RILH DI P NIL+D++F PKI+
Sbjct: 220 APPDNKDVFLGWEK--LQQIALGVAKGVEYLHLGCDQRILHFDINPHNILIDDHFVPKIT 393
Query: 220 DFGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRK 279
DFGLAK+C N S VSI RGT+GY+APEVFSR FG VSYKSD+YSYGML+LEM+GGRK
Sbjct: 394 DFGLAKLCPKNQSTVSITAARGTLGYIAPEVFSRNFGNVSYKSDIYSYGMLLLEMVGGRK 573
Query: 280 NYQTGGSCTSEMYFPDWIYKDLEQGNDLLNSLTISEEEND--MVKKITMVSLWCIQTNPL 337
N + ++ +P+WI+ +L + D+ +TI E+E D + KK+ +V LWCI+ NP+
Sbjct: 574 NTNMSAEESFQVLYPEWIH-NLLKSRDV--QVTI-EDEGDVRIAKKLAIVGLWCIEWNPI 741
Query: 338 DRPPMNKVIEMLQGPLSSVTFPPKP 362
DRP M VI+ML+G + PP P
Sbjct: 742 DRPSMKTVIQMLEGDGDKLIAPPTP 816
>TC222292 similar to UP|Q9FID5 (Q9FID5) Receptor protein kinase-like protein,
partial (16%)
Length = 732
Score = 256 bits (654), Expect = 1e-68
Identities = 118/186 (63%), Positives = 150/186 (80%)
Frame = +2
Query: 174 NTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSI 233
+ +QIAIGIARGLEYLH+GC++RILH DIKP NILLDENFCPKISDFGLAK+C SI
Sbjct: 11 DNFWQIAIGIARGLEYLHRGCNTRILHFDIKPHNILLDENFCPKISDFGLAKLCPRKGSI 190
Query: 234 VSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYF 293
+S+ RGTIGY+APEV++R FGGVS+KSDVY YGM++LEM+GGR N S TSE+YF
Sbjct: 191 ISMSDPRGTIGYVAPEVWNRNFGGVSHKSDVYRYGMMLLEMVGGRNNINAEASHTSEIYF 370
Query: 294 PDWIYKDLEQGNDLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQGPL 353
PDWIYK LEQG DL + ++ EEN++VK++ +V LWC+QT P DRP M +V++ML+G +
Sbjct: 371 PDWIYKRLEQGGDLRPNGVMAPEENEIVKRMPVVGLWCVQTFPKDRPAMTRVVDMLEGKM 550
Query: 354 SSVTFP 359
+S+ P
Sbjct: 551 NSLDIP 568
>TC209519 similar to UP|Q84K89 (Q84K89) Receptor kinase LRK10 (Receptor
kinase LRK14), partial (19%)
Length = 866
Score = 241 bits (614), Expect = 6e-64
Identities = 120/245 (48%), Positives = 160/245 (64%)
Frame = +2
Query: 137 CYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSS 196
C E K AL+YEFMP GSLDK+I+ + + ++I +GIARG+ YLHQ C
Sbjct: 2 CAEGEKHALVYEFMPNGSLDKYIFSK---EESVSLSYDKTYEICLGIARGIAYLHQDCDV 172
Query: 197 RILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFG 256
+ILH DIKP NILLD+NF PK+SDFGLAK+ + D + + G RGT GYMAPE+F + G
Sbjct: 173 QILHFDIKPHNILLDDNFIPKVSDFGLAKLYPIKDKSIILTGLRGTFGYMAPELFYKNIG 352
Query: 257 GVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYFPDWIYKDLEQGNDLLNSLTISEE 316
GVSYK+DVYS+GML++EM R+N +S+ +FP WIY + D ++ +SEE
Sbjct: 353 GVSYKADVYSFGMLLMEMGSRRRNSNPHTEHSSQHFFPFWIYDHFMEEKD-IHMEEVSEE 529
Query: 317 ENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQGPLSSVTFPPKPVLFSPKRPPLQLSN 376
+ +VKK+ +VSLWCIQ P DRP M KV+EML+G + ++ PPKPV F P +
Sbjct: 530 DKILVKKMFIVSLWCIQLKPNDRPSMKKVVEMLEGKVENIDMPPKPV-FYPHETTIDSDQ 706
Query: 377 MSSSD 381
S SD
Sbjct: 707 ASWSD 721
>TC211548 weakly similar to UP|Q7XIP7 (Q7XIP7) Receptor-like kinase
TAK33-like protein, partial (58%)
Length = 1194
Score = 239 bits (611), Expect = 1e-63
Identities = 125/304 (41%), Positives = 197/304 (64%), Gaps = 4/304 (1%)
Frame = +2
Query: 64 RYSYAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQVAVKVINES--KGNGEEFINEVAS 121
R++ +++ T+++ LG GG+GVVYK S ++G VAVKV+ S K E+F+ EV +
Sbjct: 11 RFTDQQLRIATDNYSFLLGSGGFGVVYKGSFSNGTIVAVKVLRGSSDKRIDEQFMAEVGT 190
Query: 122 ISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAI 181
I + H N+V L G+C+E + RAL+YE+M G+L+K+++ L +IA+
Sbjct: 191 IGKVHHFNLVRLYGFCFERHLRALVYEYMVNGALEKYLFHEN-----TTLSFEKLHEIAV 355
Query: 182 GIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRG 241
G ARG+ YLH+ C RI+H DIKP NILLD NFCPK++DFGLAK+C +++ +S+ G RG
Sbjct: 356 GTARGIAYLHEECQQRIIHYDIKPGNILLDRNFCPKVADFGLAKLCNRDNTHISMTGGRG 535
Query: 242 TIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYFPDWIYK-- 299
T GY APE++ V++K DVYS+GML+ E+IG R+N+ S+++FP W+++
Sbjct: 536 TPGYAAPELWLPF--PVTHKCDVYSFGMLLFEIIGRRRNHNI-NLPESQVWFPMWVWERF 706
Query: 300 DLEQGNDLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQGPLSSVTFP 359
D E DL+++ I ++ ++ ++I V+L C+Q P RP M+ V++ML+G +
Sbjct: 707 DAENVEDLISACGIEDQNREIAERIVKVALSCVQYKPEARPIMSVVVKMLEGSVE----V 874
Query: 360 PKPV 363
PKP+
Sbjct: 875 PKPL 886
>TC207391 similar to UP|Q9SWU8 (Q9SWU8) Receptor-like kinase (Fragment),
partial (29%)
Length = 963
Score = 156 bits (395), Expect(2) = 4e-61
Identities = 76/139 (54%), Positives = 95/139 (67%)
Frame = +1
Query: 115 FINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSN 174
FINEV + + H+N+V LLGYC E RAL+Y F P GSL FI+ +
Sbjct: 19 FINEVEIMGKIHHINVVRLLGYCAEGIHRALVYNFFPNGSLQSFIFPPDDKQNFLGWEK- 195
Query: 175 TLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIV 234
L IA+GIA+G+ YLHQGC+ I+H DI P N+LLD+NF PKISDFGLAK+C N S+V
Sbjct: 196 -LQNIALGIAKGIGYLHQGCNHPIIHFDINPHNVLLDDNFTPKISDFGLAKLCSKNPSLV 372
Query: 235 SIPGTRGTIGYMAPEVFSR 253
S+ RGT+GY+APEVFSR
Sbjct: 373 SMTAARGTLGYIAPEVFSR 429
Score = 97.1 bits (240), Expect(2) = 4e-61
Identities = 51/111 (45%), Positives = 71/111 (63%), Gaps = 3/111 (2%)
Frame = +2
Query: 255 FGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYFPDWIYKDLEQGNDLLNSLTIS 314
FG VSYKSD+YSYGML+LEM+GGRKN T + +PDW++ DL G+ ++
Sbjct: 434 FGNVSYKSDIYSYGMLLLEMVGGRKNVDTSSPEDFHVLYPDWMH-DLVHGDVHIH----V 598
Query: 315 EEEND--MVKKITMVSLWCIQTNPLDRPPMNKVIEMLQGPLSS-VTFPPKP 362
E+E D + +K+ +V LWCIQ PL+RP + VI+ML+ +T PP P
Sbjct: 599 EDEGDVKIARKLAIVGLWCIQWQPLNRPSIKSVIQMLESKEEDLLTVPPNP 751
>TC216543 weakly similar to UP|Q84V32 (Q84V32) Receptor kinase LRK45, partial
(23%)
Length = 603
Score = 210 bits (534), Expect = 1e-54
Identities = 98/183 (53%), Positives = 129/183 (69%)
Frame = +2
Query: 183 IARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRGT 242
+ARG+ YLH GC +ILH DIKP N LDENF PK+SDFGLAK+ +++SIV RGT
Sbjct: 2 VARGIAYLHYGCEMQILHFDIKPHNXXLDENFTPKVSDFGLAKLYPIDNSIVPRTAARGT 181
Query: 243 IGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYFPDWIYKDLE 302
IGYMAPE+F GG+S+K+DVYSYGML++EM RKN +S+++FP WIY +
Sbjct: 182 IGYMAPELFYNNIGGISHKADVYSYGMLLMEMASKRKNLNPHAERSSQLFFPFWIYNHIG 361
Query: 303 QGNDLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQGPLSSVTFPPKP 362
D + ++EEE MVKK+ +V+LWCIQ P DRP MNKV+EML+G + ++ PPKP
Sbjct: 362 DEED-IEMEDVTEEEKKMVKKMIIVALWCIQLKPNDRPSMNKVVEMLEGDIENLEIPPKP 538
Query: 363 VLF 365
L+
Sbjct: 539 TLY 547
>TC204549 similar to UP|Q9M0X5 (Q9M0X5) Receptor protein kinase-like protein,
partial (54%)
Length = 2032
Score = 207 bits (527), Expect = 8e-54
Identities = 130/368 (35%), Positives = 212/368 (57%), Gaps = 26/368 (7%)
Frame = +3
Query: 40 RKRRKHVDNNVEVFMQSYNLSI--ARRYSYAEVKRITNSFR--DKLGHGGYGVVYKASLT 95
R+ RK +V+ +Y++ + ++ ++ ++ TN F +KLG GG+G VYK +L+
Sbjct: 711 RRARKKQQGSVKEGKTAYDIPTVDSLQFDFSTIEAATNKFSADNKLGEGGFGEVYKGTLS 890
Query: 96 DGRQVAVKVINESKGNG-EEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGS 154
G+ VAVK +++S G G EEF NEV +++ H N+V LLG+C + ++ L+YE++P S
Sbjct: 891 SGQVVAVKRLSKSSGQGGEEFKNEVVVVAKLQHRNLVRLLGFCLQGEEKILVYEYVPNKS 1070
Query: 155 LDKFIYKSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENF 214
LD ++ P+ + D ++I GIARG++YLH+ RI+H D+K NILLD +
Sbjct: 1071 LDYILFD---PEKQRELDWGRRYKIIGGIARGIQYLHEDSRLRIIHRDLKASNILLDGDM 1241
Query: 215 CPKISDFGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEM 274
PKISDFG+A+I ++ + + GT GYMAPE G S KSDVYS+G+L++E+
Sbjct: 1242 NPKISDFGMARIFGVDQTQGNTSRIVGTYGYMAPEY--AMHGEFSVKSDVYSFGVLLMEI 1415
Query: 275 IGGRKN---YQTGGSCTSEMYFPDW-IYKD---LEQGNDLLNSLTISEEENDMVKKITMV 327
+ G+KN YQT G+ ++ W ++KD LE + +L S +N++++ I +
Sbjct: 1416 LSGKKNSSFYQTDGA--EDLLSYAWQLWKDGTPLELMDPILRE---SYNQNEVIRSI-HI 1577
Query: 328 SLWCIQTNPLDRPPMNKVIEMLQGPLSSVTFPPKPVLF-----SPKRP---------PLQ 373
L C+Q +P DRP M ++ ML ++ P +P F P P P+
Sbjct: 1578 GLLCVQEDPADRPTMATIVLMLDSNTVTLPTPTQPAFFVHSGTDPNMPKELPFDQSIPMS 1757
Query: 374 LSNMSSSD 381
+++MS S+
Sbjct: 1758 VNDMSISE 1781
>TC231087 similar to UP|Q39203 (Q39203) Receptor-like protein kinase
precursor, partial (30%)
Length = 826
Score = 199 bits (505), Expect = 3e-51
Identities = 111/284 (39%), Positives = 170/284 (59%), Gaps = 16/284 (5%)
Frame = +1
Query: 81 LGHGGYGVVYKASLTDGRQVAVKVINESKGNGEEFINEVASISRTSHMNIVSLLGYCYEA 140
+GHGG+G V++ L+D VAVK + G +EF EV++I H+N+V L G+C E
Sbjct: 1 VGHGGFGTVFQGELSDASVVAVKRLERPGGGEKEFRAEVSTIGNIQHVNLVRLRGFCSEN 180
Query: 141 NKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDS-NTLFQIAIGIARGLEYLHQGCSSRIL 199
+ R L+YE+M G+L+ ++ K G C S + F++A+G A+G+ YLH+ C I+
Sbjct: 181 SHRLLVYEYMQNGALNVYLRKEG------PCLSWDVRFRVAVGTAKGIAYLHEECRCCII 342
Query: 200 HLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVS 259
H DIKP+NILLD +F K+SDFGLAK+ + S V + RGT GY+APE S ++
Sbjct: 343 HCDIKPENILLDGDFTAKVSDFGLAKLIGRDFSRVLVT-MRGTWGYVAPEWISGV--AIT 513
Query: 260 YKSDVYSYGMLILEMIGGRKNYQT---------GGSCTSEM----YFPDWIYKDLEQGN- 305
K+DVYSYGM +LE+IGGR+N + GG EM +FP W + + +GN
Sbjct: 514 TKADVYSYGMTLLELIGGRRNVEAPLSAGGGGGGGESGDEMGGKWFFPPWAAQRIIEGNV 693
Query: 306 -DLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEM 348
D+++ + + +++ +V++WCIQ + RP M V++M
Sbjct: 694 SDVMDKRLGNAYNIEEARRVALVAVWCIQDDEAMRPTMGMVVKM 825
>TC209776 similar to UP|O65470 (O65470) Serine/threonine kinase-like protein
, partial (47%)
Length = 1305
Score = 198 bits (503), Expect = 5e-51
Identities = 120/331 (36%), Positives = 186/331 (55%), Gaps = 7/331 (2%)
Frame = +2
Query: 64 RYSYAEVKRITNSFRD--KLGHGGYGVVYKASLTDGRQVAVKVINESKGNG-EEFINEVA 120
R+ ++ ++ T F + KLG GG+G VYK L G++VAVK +++ G G EEF NEV
Sbjct: 41 RFDFSTIEAATQKFSEANKLGEGGFGEVYKGLLPSGQEVAVKRLSKISGQGGEEFKNEVE 220
Query: 121 SISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIA 180
+++ H N+V LLG+C E ++ L+YEF+ SLD ++ P+ D ++I
Sbjct: 221 IVAKLQHRNLVRLLGFCLEGEEKILVYEFVANKSLDYILFD---PEKQKSLDWTRRYKIV 391
Query: 181 IGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTR 240
GIARG++YLH+ +I+H D+K N+LLD + PKISDFG+A+I ++ + +
Sbjct: 392 EGIARGIQYLHEDSRLKIIHRDLKASNVLLDGDMNPKISDFGMARIFGVDQTQANTNRIV 571
Query: 241 GTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKN---YQTGGSCTSEMYFPDW- 296
GT GYM+PE G S KSDVYS+G+LILE+I G++N Y+T ++ W
Sbjct: 572 GTYGYMSPEY--AMHGEYSAKSDVYSFGVLILEIISGKRNSSFYET--DVAEDLLSYAWK 739
Query: 297 IYKDLEQGNDLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQGPLSSV 356
++KD + SL S N++++ I + L C+Q +P+DRP M V+ ML ++
Sbjct: 740 LWKDEAPLELMDQSLRESYTRNEVIRCI-HIGLLCVQEDPIDRPTMASVVLMLDSYSVTL 916
Query: 357 TFPPKPVLFSPKRPPLQLSNMSSSDWQETNS 387
P +P + R + D TNS
Sbjct: 917 QVPNQPAFYINSRTEPNMPKGLKIDQSTTNS 1009
>TC222220 weakly similar to UP|Q9ZP16 (Q9ZP16) Receptor-like protein kinase,
RLK3 precursor, partial (35%)
Length = 1304
Score = 197 bits (502), Expect = 6e-51
Identities = 122/379 (32%), Positives = 200/379 (52%), Gaps = 4/379 (1%)
Frame = +3
Query: 15 VTMFFIIFISCYFKKGIRRPQMTIFRKRRKHVDNNVEVFMQSYNLSIARRYSYAEVKRIT 74
V ++ CYF + R R+K+ E F + + ++ ++ T
Sbjct: 66 VVSIILLLCVCYF---------ILKRSRKKYNTLLRENFGEESATLESLQFGLVTIEAAT 218
Query: 75 N--SFRDKLGHGGYGVVYKASLTDGRQVAVKVINESKGNG-EEFINEVASISRTSHMNIV 131
N S+ ++G GG+GVVYK L DGR++AVK +++S G G EF NE+ I++ H N+V
Sbjct: 219 NKFSYEKRIGEGGFGVVYKGVLPDGREIAVKKLSKSSGQGANEFKNEILLIAKLQHRNLV 398
Query: 132 SLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAIGIARGLEYLH 191
+LLG+C E +++ LIYEF+ SLD F++ S + + + ++I GIA+G+ YLH
Sbjct: 399 TLLGFCLEEHEKMLIYEFVSNKSLDYFLFDSHRSKQL---NWSERYKIIEGIAQGISYLH 569
Query: 192 QGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRGTIGYMAPEVF 251
+ +++H D+KP N+LLD N PKISDFG+A+I ++ GT GYM+PE
Sbjct: 570 EHSRLKVIHRDLKPSNVLLDSNMNPKISDFGMARIVAIDQLQGKTNRIVGTYGYMSPEY- 746
Query: 252 SRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYFPDW-IYKDLEQGNDLLNS 310
G S KSDV+S+G+++LE+I ++N ++ S ++ W + D N S
Sbjct: 747 -AMHGQFSEKSDVFSFGVIVLEIISAKRNTRSVFSDHDDLLSYTWEQWMDEAPLNIFDQS 923
Query: 311 LTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQGPLSSVTFPPKPVLFSPKRP 370
+ ++ V K + L C+Q P DRP + +VI L ++ + P KP+ S
Sbjct: 924 IEAEFCDHSEVVKCIQIGLLCVQEKPDDRPKITQVISYLNSSITELPLPKKPIRQSGIVQ 1103
Query: 371 PLQLSNMSSSDWQETNSIT 389
+ + SS N ++
Sbjct: 1104KIAVGESSSGSTPSINEMS 1160
>TC220351 similar to UP|Q961E7 (Q961E7) GH28523p (CG1830-PB) , partial (11%)
Length = 1064
Score = 197 bits (502), Expect = 6e-51
Identities = 111/267 (41%), Positives = 159/267 (58%), Gaps = 4/267 (1%)
Frame = +1
Query: 100 VAVKVINESKGNGEEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFI 159
VAVK + + ++F EVA+IS T H+N+V L+G+C E R L+YEFM GSLD F+
Sbjct: 1 VAVKQLEGIEQGEKQFRMEVATISSTHHLNLVRLIGFCSEGRHRLLVYEFMKNGSLDDFL 180
Query: 160 YKSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKIS 219
+ + + F IA+G ARG+ YLH+ C I+H DIKP+NILLDEN+ K+S
Sbjct: 181 FLTEQHSGKL-LNWEYRFNIALGTARGITYLHEECRDCIVHCDIKPENILLDENYVAKVS 357
Query: 220 DFGLAKICQMND-SIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGR 278
DFGLAK+ D ++ RGT GY+APE + ++ KSDVY YGM++LE++ GR
Sbjct: 358 DFGLAKLINPKDHRHRTLTSVRGTRGYLAPEWLANL--PITSKSDVYGYGMVLLEIVSGR 531
Query: 279 KNYQTGGSCTSEMYFPDWIYKDLEQGN---DLLNSLTISEEENDMVKKITMVSLWCIQTN 335
+N+ T+ F W Y++ E+GN L L E + + V++ S WCIQ
Sbjct: 532 RNFDVSEE-TNRKKFSIWAYEEFEKGNISGILDKRLANQEVDMEQVRRAIQASFWCIQEQ 708
Query: 336 PLDRPPMNKVIEMLQGPLSSVTFPPKP 362
P RP M++V++ML+G VT P +P
Sbjct: 709 PSHRPTMSRVLQMLEG----VTEPERP 777
>TC222059 similar to GB|CAD39676.1|21741394|OSJN00121 OSJNBb0051N19.7 {Oryza
sativa (japonica cultivar-group);} , partial (30%)
Length = 510
Score = 184 bits (467), Expect = 7e-47
Identities = 89/157 (56%), Positives = 115/157 (72%)
Frame = +2
Query: 219 SDFGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGR 278
SDFGLAKIC +S++SI G RGT GY+APEVFSR FG VS+KSDVYSYGM+ILEM+G R
Sbjct: 2 SDFGLAKICTRKESMISIFGARGTAGYIAPEVFSRNFGTVSHKSDVYSYGMMILEMVGRR 181
Query: 279 KNYQTGGSCTSEMYFPDWIYKDLEQGNDLLNSLTISEEENDMVKKITMVSLWCIQTNPLD 338
KN +T +C+SE+YFPDWIY LE +L +E ++ +V+K+T+V LWCIQT+P
Sbjct: 182 KNIKTEVNCSSEIYFPDWIYNRLESNEELGLQNIRNESDDKLVRKMTIVGLWCIQTHPST 361
Query: 339 RPPMNKVIEMLQGPLSSVTFPPKPVLFSPKRPPLQLS 375
RP ++KV+EML + + PPKP L SP P+ LS
Sbjct: 362 RPAISKVLEMLGSKVELLQIPPKPFLSSPPTSPVHLS 472
>TC218138 homologue to UP|Q8GRK2 (Q8GRK2) Somatic embryogenesis receptor
kinase 1, partial (60%)
Length = 1444
Score = 181 bits (459), Expect = 6e-46
Identities = 116/304 (38%), Positives = 169/304 (55%), Gaps = 10/304 (3%)
Frame = +1
Query: 58 NLSIARRYSYAEVKRITNSFRDK--LGHGGYGVVYKASLTDGRQVAVKVINESKGNGEE- 114
+L +R+S E++ T+SF +K LG GG+G VYK L DG VAVK + E + G E
Sbjct: 100 HLGQLKRFSLRELQVATDSFSNKNILGRGGFGKVYKGRLADGSLVAVKRLKEERTPGGEL 279
Query: 115 -FINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDS 173
F EV IS H N++ L G+C +R L+Y +M GS+ + + P D
Sbjct: 280 QFQTEVEMISMAVHRNLLRLRGFCMTPTERLLVYPYMANGSVASCLRER--PPYQEPLDW 453
Query: 174 NTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSI 233
T ++A+G ARGL YLH C +I+H D+K NILLDE F + DFGLAK+ D+
Sbjct: 454 PTRKRVALGSARGLSYLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFGLAKLMDYKDTH 633
Query: 234 VSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCT-SEMY 292
V+ RGTIG++APE S G S K+DV+ YG+++LE+I G++ + ++
Sbjct: 634 VT-TAVRGTIGHIAPEYLST--GKSSEKTDVFGYGIMLLELITGQRAFDLARLANDDDVM 804
Query: 293 FPDWI-----YKDLEQGNDLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIE 347
DW+ K LE D E E V+++ V+L C Q +P+DRP M++V+
Sbjct: 805 LLDWVKGLLKEKKLEMLVDPDLQTNYIETE---VEQLIQVALLCTQGSPMDRPKMSEVVR 975
Query: 348 MLQG 351
ML+G
Sbjct: 976 MLEG 987
>TC209989 similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kinase homolog
RK20-1, partial (32%)
Length = 738
Score = 180 bits (457), Expect = 1e-45
Identities = 95/219 (43%), Positives = 141/219 (64%), Gaps = 3/219 (1%)
Frame = +3
Query: 64 RYSYAEVKRITNSFRD--KLGHGGYGVVYKASLTDGRQVAVKVINESKGNGE-EFINEVA 120
++ +A +K TN+F D KLG GG+G+VYK +L+DG+++A+K ++ + GE EF NE+
Sbjct: 96 QFEFATIKFATNNFSDANKLGQGGFGIVYKGTLSDGQEIAIKRLSINSNQGETEFKNEIL 275
Query: 121 SISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIA 180
R H N+V LLG+C+ +R LIYEF+P SLD FI+ P+ + + ++I
Sbjct: 276 LTGRLQHRNLVRLLGFCFARRERLLIYEFVPNKSLDYFIFD---PNKRVNLN*EIRYKII 446
Query: 181 IGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTR 240
GIARGL YLH+ ++H D+K NILLD PKISDFG+A++ ++N + S
Sbjct: 447 RGIARGLLYLHEDSRLNVVHRDLKTSNILLDGELNPKISDFGMARLFEINQTEASTTTIV 626
Query: 241 GTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRK 279
GT GYMAPE G S KSDV+S+G++ILE++ G++
Sbjct: 627 GTFGYMAPEYIKH--GQFSIKSDVFSFGVMILEIVCGQR 737
>TC219154 similar to PIR|T05754|T05754 S-receptor kinase M4I22.110 precursor
- Arabidopsis thaliana {Arabidopsis thaliana;} ,
partial (31%)
Length = 1363
Score = 179 bits (453), Expect = 3e-45
Identities = 120/308 (38%), Positives = 167/308 (53%), Gaps = 5/308 (1%)
Frame = +1
Query: 79 DKLGHGGYGVVYKASLTDGRQVAVKVINESKGNG-EEFINEVASISRTSHMNIVSLLGYC 137
+K+G GG+G VYK L G+++AVK ++ G G EFI EV I++ H N+V LLG C
Sbjct: 361 NKIGEGGFGPVYKGKLVGGQEIAVKRLSSLSGQGITEFITEVKLIAKLQHRNLVKLLGCC 540
Query: 138 YEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSR 197
+ ++ L+YE++ GSL+ FI+ + D F I +GIARGL YLHQ R
Sbjct: 541 IKGQEKLLVYEYVVNGSLNSFIFDQIKSKLL---DWPRRFNIILGIARGLLYLHQDSRLR 711
Query: 198 ILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAF-G 256
I+H D+K N+LLDE PKISDFG+A+ + + + GT GYMAPE AF G
Sbjct: 712 IIHRDLKASNVLLDEKLNPKISDFGMARAFGGDQTEGNTNRVVGTYGYMAPEY---AFDG 882
Query: 257 GVSYKSDVYSYGMLILEMIGGRKNYQ-TGGSCTSEMYFPDWIYKDLEQGNDLLNSLTISE 315
S KSDV+S+G+L+LE++ G KN + T + W + L++S
Sbjct: 883 NFSIKSDVFSFGILLLEIVCGIKNKSLCHENQTLNLVGYAWALWKEQNALQLIDSGIKDS 1062
Query: 316 EENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQGPLSSVTFPPKPVLFSPKR--PPLQ 373
V + VSL C+Q P DRP M VI+ML + V PK F P+R
Sbjct: 1063CVIPEVLRCIHVSLLCVQQYPEDRPTMTSVIQMLGSEMDMV--EPKEPGFFPRRILKEGN 1236
Query: 374 LSNMSSSD 381
L M+S+D
Sbjct: 1237LKEMTSND 1260
>TC217441 homologue to GB|AAK68074.1|14573459|AF384970 somatic embryogenesis
receptor-like kinase 3 {Arabidopsis thaliana;} , partial
(54%)
Length = 1008
Score = 177 bits (450), Expect = 7e-45
Identities = 114/328 (34%), Positives = 181/328 (54%), Gaps = 17/328 (5%)
Frame = +2
Query: 41 KRRKHVDNNVEVFMQS---YNLSIARRYSYAEVKRITNSFRDK--LGHGGYGVVYKASLT 95
+RRK D+ +V + +L +R+S E++ T++F +K LG GG+G VYK L
Sbjct: 38 RRRKPQDHFFDVPAEEDPEVHLGQLKRFSLRELQVATDNFSNKHILGRGGFGKVYKGRLA 217
Query: 96 DGRQVAVKVINESKGNGE--EFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKG 153
DG VAVK + E + G +F EV IS H N++ L G+C +R L+Y +M G
Sbjct: 218 DGSLVAVKRLKEERTQGG*LQFQTEVEMISMAVHRNLLRLRGFCMTPTERLLVYPYMANG 397
Query: 154 SLDKFIYKS-------GFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQ 206
S+ + + G+P+ +IA+G ARGL YLH C +I+H D+K
Sbjct: 398 SVASCLRERQESQPPLGWPERK---------RIALGSARGLAYLHDHCDPKIIHRDVKAA 550
Query: 207 NILLDENFCPKISDFGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYS 266
NILLDE F + DFGLAK+ D+ V+ RGTIG++APE S G S K+DV+
Sbjct: 551 NILLDEEFEAVVGDFGLAKLMDYKDTHVT-TAVRGTIGHIAPEYLST--GKSSEKTDVFG 721
Query: 267 YGMLILEMIGGRKNYQTGGSCT-SEMYFPDWIYKDLEQG--NDLLNSLTISEEENDMVKK 323
YG+++LE+I G++ + ++ DW+ L+ L+++ ++ V++
Sbjct: 722 YGVMLLELITGQRAFDLARLANDDDVMLLDWVKGLLKDRKLETLVDADLQGSYNDEEVEQ 901
Query: 324 ITMVSLWCIQTNPLDRPPMNKVIEMLQG 351
+ V+L C Q +P++RP M++V+ ML+G
Sbjct: 902 LIQVALLCTQGSPMERPKMSEVVRMLEG 985
>TC218530 similar to UP|Q9SGU0 (Q9SGU0) T6H22.8.1 protein, partial (25%)
Length = 1190
Score = 174 bits (440), Expect = 1e-43
Identities = 100/267 (37%), Positives = 159/267 (59%), Gaps = 3/267 (1%)
Frame = +3
Query: 99 QVAVKVINESKGNGE-EFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDK 157
++AVK ++ G+ +FI E+A+IS H N+V L G C E +KR L+YE++ SLD+
Sbjct: 3 EIAVKQLSVGSHQGKSQFITEIATISAVQHRNLVKLYGCCIEGSKRLLVYEYLENKSLDQ 182
Query: 158 FIYKSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPK 217
++ + +T + I +G+ARGL YLH+ RI+H D+K NILLD PK
Sbjct: 183 ALFGK-----CLTLNWSTRYDICLGVARGLTYLHEESRLRIVHRDVKASNILLDYELIPK 347
Query: 218 ISDFGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGG 277
ISDFGLAK+ + +S G GTIGY+APE R G ++ K+DV+S+G++ LE++ G
Sbjct: 348 ISDFGLAKLYDDKKTHIS-TGVAGTIGYLAPEYAMR--GHLTEKADVFSFGVVALELVSG 518
Query: 278 RKNYQTGGSCTSEMYFPDWIYKDLEQGNDLLNSL--TISEEENDMVKKITMVSLWCIQTN 335
R N + ++Y +W ++ L + N +++ + +SE + VK++ ++L C QT+
Sbjct: 519 RPNSDSSLE-GEKVYLLEWAWQ-LHEKNCIIDLVDDRLSEFNEEEVKRVVGIALLCTQTS 692
Query: 336 PLDRPPMNKVIEMLQGPLSSVTFPPKP 362
P RP M++V+ ML G + T KP
Sbjct: 693 PTLRPSMSRVVAMLSGDIEVSTVTSKP 773
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.138 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,533,575
Number of Sequences: 63676
Number of extensions: 266152
Number of successful extensions: 3396
Number of sequences better than 10.0: 942
Number of HSP's better than 10.0 without gapping: 2520
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2567
length of query: 459
length of database: 12,639,632
effective HSP length: 101
effective length of query: 358
effective length of database: 6,208,356
effective search space: 2222591448
effective search space used: 2222591448
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC147875.5


