

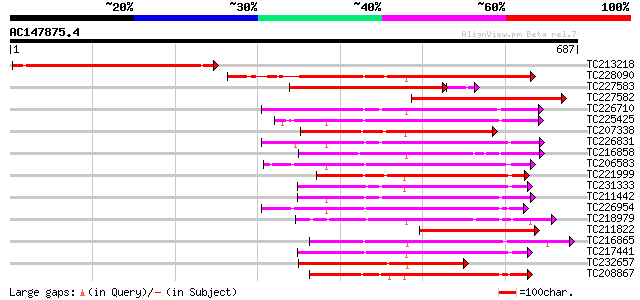
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147875.4 + phase: 0
(687 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC213218 348 3e-96
TC228090 similar to UP|Q6QLL5 (Q6QLL5) WAK-like kinase, partial ... 286 3e-77
TC227583 similar to UP|Q9LPQ6 (Q9LPQ6) F15H18.11, partial (14%) 268 7e-74
TC227582 similar to UP|Q941D0 (Q941D0) At1g25390/F2J7_14, partia... 251 1e-66
TC226710 similar to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-l... 218 9e-57
TC225425 similar to GB|AAN31120.1|23506217|AY149966 At2g17220/T2... 204 1e-52
TC207338 similar to UP|Q6K270 (Q6K270) Nodulation receptor kinas... 202 3e-52
TC226831 similar to PIR|T52285|T52285 serine/threonine-specific ... 202 5e-52
TC216858 homologue to UP|Q76FZ8 (Q76FZ8) Brassinosteroid recepto... 200 2e-51
TC206583 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B ... 198 6e-51
TC221999 homologue to UP|Q9LK35 (Q9LK35) Receptor-protein kinase... 198 8e-51
TC231333 similar to UP|Q8L741 (Q8L741) AT3g13690/MMM17_12, parti... 198 8e-51
TC211442 similar to UP|Q84QD9 (Q84QD9) Avr9/Cf-9 rapidly elicite... 196 2e-50
TC226954 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B ... 195 6e-50
TC218979 similar to UP|Q9FUU9 (Q9FUU9) Leaf senescence-associate... 195 6e-50
TC211822 weakly similar to UP|Q9LPQ6 (Q9LPQ6) F15H18.11, partial... 194 1e-49
TC216865 similar to UP|Q9LPG0 (Q9LPG0) T3F20.24 protein, partial... 193 2e-49
TC217441 homologue to GB|AAK68074.1|14573459|AF384970 somatic em... 192 5e-49
TC232657 weakly similar to UP|O22580 (O22580) Receptor-like seri... 191 7e-49
TC208867 similar to UP|Q9LXT2 (Q9LXT2) Serine/threonine-specific... 191 7e-49
>TC213218
Length = 835
Score = 348 bits (894), Expect = 3e-96
Identities = 171/251 (68%), Positives = 198/251 (78%), Gaps = 1/251 (0%)
Frame = +1
Query: 4 LSLSSPIITFIIFFIFYLRHTTSLPSHASLSSCNNTTFNCGHITKLSYPFTGGDRPSYCG 63
LSLS+ T +IF YL HTTSLP HA+LSSC+ T+FNCG IT LSYPFTGGDRPS+CG
Sbjct: 67 LSLSTITATILIF---YLHHTTSLPPHATLSSCHVTSFNCGSITILSYPFTGGDRPSFCG 237
Query: 64 PPQFHLNCKNNV-PELNISSVSYRVLQVNSVTHSLTLARLDLWNETCTHHYVNSTFGGTS 122
PPQF LNC+N V ELNISSVSYRV+ ++S H+LTLARLDLWNETCT YVNSTF G
Sbjct: 238 PPQFLLNCRNGVVAELNISSVSYRVIDIDSEDHTLTLARLDLWNETCTDVYVNSTFDGPV 417
Query: 123 FSNGLGNSKITLFYGCQPTSLFTEKPHNLFYCDSNGYKNNSYTLIGPFPLDPVLKFVQCD 182
FS G GN +TLFY C+PTS E P NLF C SNG KNNSY+L+GPFPLDP+L+ V+CD
Sbjct: 418 FSYGSGNQNLTLFYECKPTSRIIETPENLFNCWSNGDKNNSYSLVGPFPLDPILEVVECD 597
Query: 183 YGVGVPILEEQANRFAGNRSLLREVLMEGFNVNYNNPFENDCLECISSGGQQCGFDSDEN 242
V VPIL+ QA+R NRSLL EVLM+GFNVNY NP+E++C EC+ SGG CGFDSD +
Sbjct: 598 EHVKVPILKVQADRLVENRSLLGEVLMKGFNVNYMNPYESECFECLDSGG-VCGFDSDND 774
Query: 243 EHICICGNGLC 253
EHICICG+ LC
Sbjct: 775 EHICICGDHLC 807
>TC228090 similar to UP|Q6QLL5 (Q6QLL5) WAK-like kinase, partial (51%)
Length = 1533
Score = 286 bits (731), Expect = 3e-77
Identities = 163/379 (43%), Positives = 233/379 (61%), Gaps = 5/379 (1%)
Frame = +1
Query: 264 LIGGVVGGVAALCLLGFVACFVVRRRRKNAKKPISNDLYMPPSSTTSGTNTGTLTSTTNS 323
LIGG V GV+ + LG + CF RR +K ++N + LT T
Sbjct: 34 LIGGFVVGVSLMVTLGSLCCFYRRR----SKLRVTN------------STKRRLTEATGK 165
Query: 324 SQSIPSYPSSKTSTMPKSFYFGVQVFTYEELEEATNNFHTSKELGEGGFGTVYKGDLKDG 383
+ S+P YP Y+++E+ATN+F + LG G +GTVY G L +
Sbjct: 166 N-SVPIYP-------------------YKDIEKATNSFSEKQRLGTGAYGTVYAGKLYNN 285
Query: 384 RVVAVKRHYESNFKRVAQFMNEVEILARLRHKNLVTLYGCTSKHSRELLLVYEYISNGTV 443
VA+KR + + Q MNE+++L+ + H NLV L GC+ ++ E +LVYE++ NGT+
Sbjct: 286 EWVAIKRIKHRDTDSIEQVMNEIKLLSSVSHTNLVRLLGCSIEYG-EQILVYEFMPNGTL 462
Query: 444 ADHLHGDRSSSCLLPWSVRLDIALETAEALAYLHAS---DVMHRDVKSNNILLDEKFHVK 500
+ HL +R S LPW +RL IA ETA+A+AYLH++ + HRD+KS+NILLD F K
Sbjct: 463 SQHLQKERGSG--LPWPIRLTIATETAQAIAYLHSAICPPIYHRDIKSSNILLDYNFRSK 636
Query: 501 VADFGLSRLFPNDVTHVSTAPQGTPGYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQA 560
VADFGLSRL +++H+ST PQGTPGYVDP+Y+Q + L+DKSDVYS GVVLVE+I+ +
Sbjct: 637 VADFGLSRLGMTEISHISTTPQGTPGYVDPQYHQDFHLSDKSDVYSLGVVLVEIITGQKV 816
Query: 561 VDITRHRNDVNLANMAVNKIQSQELYDLVDPNLGYEKDNSVKRMTT--AVAELAFRCLQQ 618
VD +R N+VNLA++A +KI L +++DP L E + +++ VAELAFRC+
Sbjct: 817 VDFSRPHNEVNLASLAADKIGKGLLNEIIDPFLEPEVRSDAWTLSSIHKVAELAFRCIAF 996
Query: 619 QRDLRPSMDEIVEVLRAIK 637
RD+RPSM E+ L ++
Sbjct: 997 HRDMRPSMTEVASELEQLR 1053
>TC227583 similar to UP|Q9LPQ6 (Q9LPQ6) F15H18.11, partial (14%)
Length = 838
Score = 268 bits (684), Expect(2) = 7e-74
Identities = 132/193 (68%), Positives = 158/193 (81%), Gaps = 2/193 (1%)
Frame = +1
Query: 340 KSFYFGVQVFTYEELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRV 399
+SF GV +F Y+ELEEATN F +SKELGEGGFGTVY G L+DGR VAVKR YE+NFKRV
Sbjct: 13 QSFVPGVPLFLYDELEEATNYFDSSKELGEGGFGTVYFGKLRDGRSVAVKRLYENNFKRV 192
Query: 400 AQFMNEVEILARLRHKNLVTLYGCTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPW 459
AQFMNE++ILA+L H NLVTL+GCTS+H+RELLLVYEYI NGT+ADHLHG RS LPW
Sbjct: 193 AQFMNEIKILAKLDHPNLVTLFGCTSRHTRELLLVYEYIPNGTIADHLHGQRSKPGKLPW 372
Query: 460 SVRLDIALETAEALAYLHASDVMHRDVKSNNILLDEKFHVKVADFGLSRLFPNDVTHV-- 517
+R++IA+ETA AL +LH D++HRDVK+NNILLD F VKVADFGLSRLFP+ +
Sbjct: 373 HIRMNIAVETASALKFLHQKDIIHRDVKTNNILLDNNFCVKVADFGLSRLFPDPCHSMFS 552
Query: 518 STAPQGTPGYVDP 530
++ +G PGYVDP
Sbjct: 553 DSSHKGLPGYVDP 591
Score = 28.9 bits (63), Expect(2) = 7e-74
Identities = 17/40 (42%), Positives = 23/40 (57%)
Frame = +2
Query: 530 PEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRND 569
PEY+QCYQLT+K + + S AVDITR + +
Sbjct: 590 PEYHQCYQLTNKRSL*LWSXAG*P-ESPXPAVDITRXKQN 706
>TC227582 similar to UP|Q941D0 (Q941D0) At1g25390/F2J7_14, partial (34%)
Length = 861
Score = 251 bits (640), Expect = 1e-66
Identities = 121/188 (64%), Positives = 153/188 (81%)
Frame = +3
Query: 487 KSNNILLDEKFHVKVADFGLSRLFPNDVTHVSTAPQGTPGYVDPEYYQCYQLTDKSDVYS 546
K+NNILLD F VKVADFGLSRLFP+ VTHVSTAPQGTPGYVDPEY+QCYQLT +SDVYS
Sbjct: 6 KTNNILLDNNFCVKVADFGLSRLFPDHVTHVSTAPQGTPGYVDPEYHQCYQLTKQSDVYS 185
Query: 547 FGVVLVELISSLQAVDITRHRNDVNLANMAVNKIQSQELYDLVDPNLGYEKDNSVKRMTT 606
FGVVLVELISSL AVDITRHR ++ L++MA+NKI S+ L++LVDP+LG+E + V++M
Sbjct: 186 FGVVLVELISSLPAVDITRHRQEIKLSDMAINKIHSEALHELVDPSLGFESNFKVRKMIN 365
Query: 607 AVAELAFRCLQQQRDLRPSMDEIVEVLRAIKSDEPETQESKVLDVVVRTDELVLLKKGPY 666
AVAELAF+CLQ +++RPSM+E+ + L+ I+SD + +V+D+ D++VLLK P
Sbjct: 366 AVAELAFQCLQSSKEMRPSMEEVADTLKDIQSDGKHKSQPEVMDITSTADDVVLLKDDPP 545
Query: 667 PTSPDSVA 674
P SPDS A
Sbjct: 546 PPSPDSNA 569
>TC226710 similar to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-like protein,
partial (66%)
Length = 1737
Score = 218 bits (554), Expect = 9e-57
Identities = 135/347 (38%), Positives = 204/347 (57%), Gaps = 6/347 (1%)
Frame = +3
Query: 306 SSTTSGTNTGTLTSTTNSSQSIPSYPSSKTSTMPKSFYFGV-QVFTYEELEEATNNFHTS 364
S T+ G + S +S S S ++ T + S + + F++ E++ ATNNF +
Sbjct: 267 SGTSEGPSGWLPLSLYGNSHSAASAKTNTTGSYASSLPSNLCRHFSFAEIKAATNNFDEA 446
Query: 365 KELGEGGFGTVYKGDLKDGRV-VAVKRHYESNFKRVAQFMNEVEILARLRHKNLVTLYGC 423
LG GGFG VYKG++ G VA+KR + + V +F E+E+L++LRH++LV+L G
Sbjct: 447 LLLGVGGFGKVYKGEIDGGTTKVAIKRGNPLSEQGVHEFQTEIEMLSKLRHRHLVSLIGY 626
Query: 424 TSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIALETAEALAYLHAS---D 480
+++ E++LVY+Y++ GT+ +HL+ ++ PW RL+I + A L YLH
Sbjct: 627 CEENT-EMILVYDYMAYGTLREHLY--KTQKPPRPWKQRLEICIGAARGLHYLHTGAKHT 797
Query: 481 VMHRDVKSNNILLDEKFHVKVADFGLSRLFPN-DVTHVSTAPQGTPGYVDPEYYQCYQLT 539
++HRDVK+ NILLDEK+ KV+DFGLS+ P D THVST +G+ GY+DPEY++ QLT
Sbjct: 798 IIHRDVKTTNILLDEKWVAKVSDFGLSKTGPTLDNTHVSTVVKGSFGYLDPEYFRRQQLT 977
Query: 540 DKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKIQSQELYDLVDPNLGYEKDN 599
DKSDVYSFGVVL E++ + A++ T + V+LA A + Q L ++DP Y K
Sbjct: 978 DKSDVYSFGVVLFEVLCARPALNPTLAKEQVSLAEWAAHCYQKGILDSIIDP---YLKGK 1148
Query: 600 SVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVLRAIKSDEPETQES 646
AE A +C+ Q RPSM +++ L + +ES
Sbjct: 1149IAPECFKKFAETAMKCVADQGIDRPSMGDVLWNLEFALQLQESAEES 1289
>TC225425 similar to GB|AAN31120.1|23506217|AY149966 At2g17220/T23A1.8
{Arabidopsis thaliana;} , partial (80%)
Length = 1386
Score = 204 bits (518), Expect = 1e-52
Identities = 129/341 (37%), Positives = 196/341 (56%), Gaps = 15/341 (4%)
Frame = +1
Query: 322 NSSQSIPS----YPSSKTSTMPKSFYFGVQVFTYEELEEATNNFHTSKELGEGGFGTVYK 377
NS S+ S YP+ + +P S +++FT+ EL+ AT NF LGEGGFG V+K
Sbjct: 34 NSKFSVSSGDQPYPNGQI--LPTS---NLRIFTFAELKAATRNFKADTVLGEGGFGKVFK 198
Query: 378 GDLKD--------GRVVAVKRHYESNFKRVAQFMNEVEILARLRHKNLVTLYGCTSKHSR 429
G L++ G V+AVK+ + + + ++ +EV L RL H NLV L G + S
Sbjct: 199 GWLEEKATSKGGSGTVIAVKKLNSESLQGLEEWQSEVNFLGRLSHTNLVKLLGYCLEES- 375
Query: 430 ELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIALETAEALAYLHASD-VMHRDVKS 488
ELLLVYE++ G++ +HL G S+ LPW +RL IA+ A LA+LH S+ V++RD K+
Sbjct: 376 ELLLVYEFMQKGSLENHLFGRGSAVQPLPWDIRLKIAIGAARGLAFLHTSEKVIYRDFKA 555
Query: 489 NNILLDEKFHVKVADFGLSRLFPN-DVTHVSTAPQGTPGYVDPEYYQCYQLTDKSDVYSF 547
+NILLD ++ K++DFGL++L P+ +HV+T GT GY PEY L KSDVY F
Sbjct: 556 SNILLDGSYNAKISDFGLAKLGPSASQSHVTTRVMGTHGYAAPEYVATGHLYVKSDVYGF 735
Query: 548 GVVLVELISSLQAVDITRHRNDVNLANMAVNKIQS-QELYDLVDPNLGYEKDNSVKRMTT 606
GVVLVE+++ L+A+D R L + ++L ++D L + +
Sbjct: 736 GVVLVEILTGLRALDSNRPSGQHKLTEWVKPYLHDRRKLKGIMDSRL---EGKFPSKAAF 906
Query: 607 AVAELAFRCLQQQRDLRPSMDEIVEVLRAIKSDEPETQESK 647
+A+L+ +CL + RPSM +++E L I++ + E K
Sbjct: 907 RIAQLSMKCLASEPKHRPSMKDVLENLERIQAANEKPVEPK 1029
>TC207338 similar to UP|Q6K270 (Q6K270) Nodulation receptor kinase-like
protein, partial (46%)
Length = 765
Score = 202 bits (515), Expect = 3e-52
Identities = 110/245 (44%), Positives = 159/245 (64%), Gaps = 6/245 (2%)
Frame = +1
Query: 353 ELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRV-AQFMNEVEILAR 411
++ AT NF + ++GEGGFGTVYK L+DG VVAVKR + +F + +F +E+E+LA+
Sbjct: 28 QVTRATQNFSETLQIGEGGFGTVYKAKLEDGLVVAVKRAKKEHFDSLRTEFSSEIELLAK 207
Query: 412 LRHKNLVTLYGCTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIALETAE 471
+ H+NLV L G K E LL+ E++ NGT+ +HL G R +L ++ RL+IA++ A
Sbjct: 208 IDHRNLVKLLGYIDK-GNERLLITEFVPNGTLREHLDGMRGK--ILDFNQRLEIAIDVAH 378
Query: 472 ALAYLHA---SDVMHRDVKSNNILLDEKFHVKVADFGLSRLFP--NDVTHVSTAPQGTPG 526
L YLH ++HRDVKS+NILL E KVADFG +RL P D TH+ST +GT G
Sbjct: 379 GLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFARLGPVNTDQTHISTKVKGTVG 558
Query: 527 YVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKIQSQELY 586
Y+DPEY + YQLT KSDVYSFG++L+E++++ + V++ + + A K +
Sbjct: 559 YLDPEYMKTYQLTPKSDVYSFGILLLEIVTARRPVELKKTVAERVTLRWAFRKYNEGSVV 738
Query: 587 DLVDP 591
+L DP
Sbjct: 739 ELGDP 753
>TC226831 similar to PIR|T52285|T52285 serine/threonine-specific protein
kinase APK2a [imported] - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (82%)
Length = 1693
Score = 202 bits (513), Expect = 5e-52
Identities = 139/366 (37%), Positives = 204/366 (54%), Gaps = 23/366 (6%)
Frame = +1
Query: 306 SSTTSGTNTGTLTSTTNSSQSIPSYP-SSKTSTMPKSFYFG-------VQVFTYEELEEA 357
SS T + T S+ S+ SI SY +S S +P G ++ FT+ EL+ A
Sbjct: 286 SSRTPSGISKTSPSSVPSNLSILSYSEASDFSNLPTPRSEGEILSSPNLKAFTFNELKNA 465
Query: 358 TNNFHTSKELGEGGFGTVYKGDLKD----------GRVVAVKRHYESNFKRVAQFMNEVE 407
T NF LGEGGFG VYKG + + G VVAVK+ + +++ EV+
Sbjct: 466 TRNFRPDSLLGEGGFGYVYKGWIDEHTFTASKPGSGMVVAVKKLKPEGLQGHKEWLTEVD 645
Query: 408 ILARLRHKNLVTLYG-CTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIA 466
L +L H+NLV L G C +R LLVYE++S G++ +HL R L WSVR+ +A
Sbjct: 646 YLGQLHHQNLVKLIGYCADGENR--LLVYEFMSKGSLENHLF--RRGPQPLSWSVRMKVA 813
Query: 467 LETAEALAYLH--ASDVMHRDVKSNNILLDEKFHVKVADFGLSRLFPN-DVTHVSTAPQG 523
+ A L++LH S V++RD K++NILLD +F+ K++DFGL++ P D THVST G
Sbjct: 814 IGAARGLSFLHNAKSQVIYRDFKASNILLDAEFNAKLSDFGLAKAGPTGDRTHVSTQVMG 993
Query: 524 TPGYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKI-QS 582
T GY PEY +LT KSDVYSFGVVL+EL+S +AVD ++ + NL A +
Sbjct: 994 TQGYAAPEYVATGRLTAKSDVYSFGVVLLELLSGRRAVDRSKAGVEQNLVEWAKPYLGDK 1173
Query: 583 QELYDLVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVLRAIKSDEPE 642
+ L+ ++D LG + M A LA +CL ++ RP + E+++ L I + +
Sbjct: 1174RRLFRIMDTKLGGQYPQKGAYM---AATLALKCLNREAKGRPPITEVLQTLEQIAASKTA 1344
Query: 643 TQESKV 648
+ S++
Sbjct: 1345GRNSQL 1362
>TC216858 homologue to UP|Q76FZ8 (Q76FZ8) Brassinosteroid receptor, partial
(29%)
Length = 1320
Score = 200 bits (508), Expect = 2e-51
Identities = 113/303 (37%), Positives = 180/303 (59%), Gaps = 4/303 (1%)
Frame = +1
Query: 350 TYEELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNEVEIL 409
T+ +L +ATN FH +G GGFG VYK LKDG VVA+K+ + + +F E+E +
Sbjct: 49 TFADLLDATNGFHNDSLIGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQGDREFTAEMETI 228
Query: 410 ARLRHKNLVTLYGCTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIALET 469
+++H+NLV L G K E LLVYEY+ G++ D LH + + L W++R IA+
Sbjct: 229 GKIKHRNLVPLLG-YCKVGEERLLVYEYMKYGSLEDVLHDQKKAGIKLNWAIRRKIAIGA 405
Query: 470 AEALAYLHAS---DVMHRDVKSNNILLDEKFHVKVADFGLSRLFPNDVTHVSTAP-QGTP 525
A LA+LH + ++HRD+KS+N+LLDE +V+DFG++RL TH+S + GTP
Sbjct: 406 ARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTLAGTP 585
Query: 526 GYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKIQSQEL 585
GYV PEYYQ ++ + K DVYS+GVVL+EL++ + D + D NL V + ++
Sbjct: 586 GYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKRPTD-SADFGDNNLVGW-VKQHAKLKI 759
Query: 586 YDLVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVLRAIKSDEPETQE 645
D+ DP L ++D +++ ++A CL + RP+M +++ + + I++ +
Sbjct: 760 SDIFDPEL-MKEDPNLEMELLQHLKIAVSCLDDRPWRRPTMIQVMAMFKEIQAGSGIDSQ 936
Query: 646 SKV 648
S +
Sbjct: 937 STI 945
>TC206583 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B , partial
(84%)
Length = 1629
Score = 198 bits (504), Expect = 6e-51
Identities = 132/347 (38%), Positives = 196/347 (56%), Gaps = 17/347 (4%)
Frame = +1
Query: 308 TTSGTNTGTLTSTTNSSQSIPSYPSSKTSTMPKSFYFGVQVFTYEELEEATNNFHTSKEL 367
+T G + G+ T+ S+ SIP P S+ + S ++ FT EL+ AT NF L
Sbjct: 133 STDGNDLGS-TNDKVSANSIPQTPRSEGEILQSS---NLKSFTLSELKTATRNFRPDSVL 300
Query: 368 GEGGFGTVYKGDLKD----------GRVVAVKRHYESNFKRVAQFMNEVEILARLRHKNL 417
GEGGFG+V+KG + + G V+AVKR + +++ EV L + H +L
Sbjct: 301 GEGGFGSVFKGWIDENSLTATKPGTGIVIAVKRLNQDGIPGHREWLAEVNYLGQHFHSHL 480
Query: 418 VTLYG-CTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIALETAEALAYL 476
V L G C R LLVYE++ G+ +HL S L WS+RL +AL+ A+ LA+L
Sbjct: 481 VRLIGFCLEDEHR--LLVYEFMPRGSWENHLFRRGSYFQPLSWSLRLKVALDAAKGLAFL 654
Query: 477 HASD--VMHRDVKSNNILLDEKFHVKVADFGLSRLFPN-DVTHVSTAPQGTPGYVDPEYY 533
H+++ V++RD K++N+LLD K++ K++DFGL++ P D +HVST GT GY PEY
Sbjct: 655 HSAEAKVIYRDFKTSNVLLDSKYNAKLSDFGLAKDGPTGDKSHVSTRVMGTYGYAAPEYL 834
Query: 534 QCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVN-KIQSQELYDLVDPN 592
LT KSDVYSFGVVL+E++S +AVD R NL A ++++ ++D
Sbjct: 835 ATGHLTAKSDVYSFGVVLLEMLSGKRAVDKNRPSGQHNLVEWAKPFMANKRKIFRVLDTR 1014
Query: 593 L--GYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVLRAIK 637
L Y D++ K +A LA RCL + RP+MD++V L ++
Sbjct: 1015LQGQYSTDDAYK-----LATLALRCLSIESKFRPNMDQVVTTLEQLQ 1140
>TC221999 homologue to UP|Q9LK35 (Q9LK35) Receptor-protein kinase-like
protein, partial (38%)
Length = 1357
Score = 198 bits (503), Expect = 8e-51
Identities = 117/264 (44%), Positives = 170/264 (64%), Gaps = 5/264 (1%)
Frame = +1
Query: 372 FGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNEVEILARLRHKNLVTLYGCTSKHSREL 431
FG VYKG L+DG VAVKR + + +A+F E+E+L++LRH++LV+L G + S E+
Sbjct: 1 FGRVYKGTLEDGTNVAVKRGNPRSEQGLAEFRTEIEMLSKLRHRHLVSLIGYCDERS-EM 177
Query: 432 LLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIALETAEALAYLH---ASDVMHRDVKS 488
+LVYEY++NG + HL+G L W RL+I + A L YLH + ++HRDVK+
Sbjct: 178 ILVYEYMANGPLRSHLYGTDLPP--LSWKQRLEICIGAARGLHYLHTGASQSIIHRDVKT 351
Query: 489 NNILLDEKFHVKVADFGLSRLFPN-DVTHVSTAPQGTPGYVDPEYYQCYQLTDKSDVYSF 547
NIL+D+ F KVADFGLS+ P D THVSTA +G+ GY+DPEY++ QLT+KSDVYSF
Sbjct: 352 TNILVDDNFVAKVADFGLSKTGPALDQTHVSTAVKGSFGYLDPEYFRRQQLTEKSDVYSF 531
Query: 548 GVVLVELISSLQAVDITRHRNDVNLANMAVNKIQSQELYDLVDPNL-GYEKDNSVKRMTT 606
GVVL+E++ + A++ R VN+A A++ + L ++D NL G S+K+
Sbjct: 532 GVVLMEVLCTRPALNPVLPREQVNIAEWAMSWQKKGMLDQIMDQNLVGKVNPASLKKF-- 705
Query: 607 AVAELAFRCLQQQRDLRPSMDEIV 630
E A +CL + RPSM +++
Sbjct: 706 --GETAEKCLAEYGVDRPSMGDVL 771
>TC231333 similar to UP|Q8L741 (Q8L741) AT3g13690/MMM17_12, partial (38%)
Length = 1009
Score = 198 bits (503), Expect = 8e-51
Identities = 117/290 (40%), Positives = 172/290 (58%), Gaps = 5/290 (1%)
Frame = +3
Query: 349 FTYEELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNEVEI 408
FT+ EL+ AT F + L EGGFG+V++G L DG+V+AVK++ ++ + +F +EVE+
Sbjct: 6 FTFSELQLATGGFSQANFLAEGGFGSVHRGVLPDGQVIAVKQYKLASTQGDKEFCSEVEV 185
Query: 409 LARLRHKNLVTLYGCTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIALE 468
L+ +H+N+V L G R LL VYEYI NG++ H++ R +L WS R IA+
Sbjct: 186 LSCAQHRNVVMLIGFCVDDGRRLL-VYEYICNGSLDSHIY--RRKQNVLEWSARQKIAVG 356
Query: 469 TAEALAYLH----ASDVMHRDVKSNNILLDEKFHVKVADFGLSRLFPNDVTHVSTAPQGT 524
A L YLH ++HRD++ NNILL F V DFGL+R P+ V T GT
Sbjct: 357 AARGLRYLHEECRVGCIVHRDMRPNNILLTHDFEALVGDFGLARWQPDGDMGVETRVIGT 536
Query: 525 PGYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKIQSQE 584
GY+ PEY Q Q+T+K+DVYSFG+VL+EL++ +AVDI R + L+ A ++ Q
Sbjct: 537 FGYLAPEYAQSGQITEKADVYSFGIVLLELVTGRKAVDINRPKGQQCLSEWARPLLEKQA 716
Query: 585 LYDLVDPNL-GYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVL 633
Y L+DP+L D V RM + + C+ + LRP M +++ +L
Sbjct: 717 TYKLIDPSLRNCYVDQEVYRM----LKCSSLCIGRDPHLRPRMSQVLRML 854
>TC211442 similar to UP|Q84QD9 (Q84QD9) Avr9/Cf-9 rapidly elicited protein
264, partial (73%)
Length = 1047
Score = 196 bits (499), Expect = 2e-50
Identities = 123/301 (40%), Positives = 178/301 (58%), Gaps = 12/301 (3%)
Frame = +2
Query: 349 FTYEELEEATNNFHTSKELGEGGFGTVYKGDLKD-------GRVVAVKRHYESNFKRVAQ 401
FT EEL EATN+F S LGEGGFG VYKG + D + VAVKR + +
Sbjct: 35 FTLEELREATNSFSWSNMLGEGGFGPVYKGFVDDKLRSGLKAQTVAVKRLDLDGLQGHRE 214
Query: 402 FMNEVEILARLRHKNLVTLYG-CTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWS 460
++ E+ L +LRH +LV L G C R LL+YEY+ G++ + L R S +PWS
Sbjct: 215 WLAEIIFLGQLRHPHLVKLIGYCYEDEHR--LLMYEYMPRGSLENQLF--RRYSAAMPWS 382
Query: 461 VRLDIALETAEALAYLHASD--VMHRDVKSNNILLDEKFHVKVADFGLSRLFPN-DVTHV 517
R+ IAL A+ LA+LH +D V++RD K++NILLD F K++DFGL++ P + THV
Sbjct: 383 TRMKIALGAAKGLAFLHEADKPVIYRDFKASNILLDSDFTAKLSDFGLAKDGPEGEDTHV 562
Query: 518 STAPQGTPGYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAV 577
+T GT GY PEY LT KSDVYS+GVVL+EL++ + VD +R +L A
Sbjct: 563 TTRIMGTQGYAAPEYIMTGHLTTKSDVYSYGVVLLELLTGRRVVDKSRSNEGKSLVEWAR 742
Query: 578 NKIQSQ-ELYDLVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVLRAI 636
++ Q ++Y+++D L + + VA LAF+CL + RP+M ++++VL +
Sbjct: 743 PLLRDQKKVYNIIDRRL---EGQFPMKGAMKVAMLAFKCLSHHPNARPTMSDVIKVLEPL 913
Query: 637 K 637
+
Sbjct: 914 Q 916
>TC226954 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B , partial
(80%)
Length = 1175
Score = 195 bits (495), Expect = 6e-50
Identities = 128/338 (37%), Positives = 192/338 (55%), Gaps = 15/338 (4%)
Frame = +2
Query: 306 SSTTSGTNTGTLTSTTNSSQSIPSYPSSKTSTMPKSFYFGVQVFTYEELEEATNNFHTSK 365
S + S + +++ +SS SIP S+ + S ++ F+Y EL AT NF
Sbjct: 185 SRSVSRSGHDISSNSRSSSASIPVTSRSEGEILQSS---NLKSFSYHELRAATRNFRPDS 355
Query: 366 ELGEGGFGTVYKGDLKD----------GRVVAVKRHYESNFKRVAQFMNEVEILARLRHK 415
LGEGGFG+V+KG + + G++VAVK+ + + +++ E+ L +L+H
Sbjct: 356 VLGEGGFGSVFKGWIDEHSLAATKPGIGKIVAVKKLNQDGLQGHREWLAEINYLGQLQHP 535
Query: 416 NLVTLYG-CTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIALETAEALA 474
NLV L G C R LLVYE++ G++ +HL S WS+R+ IAL A+ LA
Sbjct: 536 NLVKLIGYCFEDEHR--LLVYEFMPKGSMENHLFRRGSYFQPFSWSLRMKIALGAAKGLA 709
Query: 475 YLHASD--VMHRDVKSNNILLDEKFHVKVADFGLSRLFPN-DVTHVSTAPQGTPGYVDPE 531
+LH+++ V++RD K++NILLD ++ K++DFGL+R P D +HVST GT GY PE
Sbjct: 710 FLHSTEHKVIYRDFKTSNILLDTNYNAKLSDFGLARDGPTGDKSHVSTRVMGTRGYAAPE 889
Query: 532 YYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKIQS-QELYDLVD 590
Y LT KSDVYSFGVVL+E+IS +A+D + + NL A + + + ++ ++D
Sbjct: 890 YLATGHLTTKSDVYSFGVVLLEMISGRRAIDKNQPTGEHNLVEWAKPYLSNKRRVFRVMD 1069
Query: 591 PNLGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDE 628
P L E S R A A LA +C + RP+MDE
Sbjct: 1070PRL--EGQYSQNR-AQAAAALAMQCFSVEPKCRPNMDE 1174
>TC218979 similar to UP|Q9FUU9 (Q9FUU9) Leaf senescence-associated
receptor-like protein kinase, partial (34%)
Length = 1343
Score = 195 bits (495), Expect = 6e-50
Identities = 125/329 (37%), Positives = 189/329 (56%), Gaps = 13/329 (3%)
Frame = +3
Query: 347 QVFTYEELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNEV 406
Q+++Y ++ + TNNF+T +G+GGFGTVY G + D V AVK S+ QF EV
Sbjct: 153 QIYSYSDVLKITNNFNTI--IGKGGFGTVYLGYIDDSPV-AVKVLSPSSVNGFQQFQAEV 323
Query: 407 EILARLRHKNLVTLYGCTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIA 466
++L ++ HKNL +L G ++ + + L +YEY++NG + +HL G S S L W RL IA
Sbjct: 324 KLLVKVHHKNLTSLIGYCNEGTNKAL-IYEYMANGNLQEHLSGKHSKSTFLSWEDRLRIA 500
Query: 467 LETAEALAYLHAS---DVMHRDVKSNNILLDEKFHVKVADFGLSRLFPND-VTHVSTAPQ 522
++ A L YL ++HRDVKS NILL+E F K++DFGLS+ P D +HVST
Sbjct: 501 VDAALGLEYLQNGCKPPIIHRDVKSTNILLNEHFQAKLSDFGLSKAIPTDGESHVSTVLA 680
Query: 523 GTPGYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKIQS 582
GTPGY+DP + +LT KSDVYSFGVVL+E+I++ ++ R++ ++ + I+
Sbjct: 681 GTPGYLDPHCHISSRLTQKSDVYSFGVVLLEIITNQPVME--RNQEKGHIRERVRSLIEK 854
Query: 583 QELYDLVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEI---------VEVL 633
++ +VD L E D + A+ E+A C+ Q + RP M EI +E+
Sbjct: 855 GDIRAIVDSRL--EGDYDINSAWKAL-EIAMACVSQNPNERPIMSEIAIELKETLAIEIA 1025
Query: 634 RAIKSDEPETQESKVLDVVVRTDELVLLK 662
RA D + + V V T+ + L +
Sbjct: 1026RAKHCDANPRYLVEAVSVNVDTEFMPLAR 1112
>TC211822 weakly similar to UP|Q9LPQ6 (Q9LPQ6) F15H18.11, partial (12%)
Length = 465
Score = 194 bits (493), Expect = 1e-49
Identities = 95/146 (65%), Positives = 118/146 (80%)
Frame = +2
Query: 497 FHVKVADFGLSRLFPNDVTHVSTAPQGTPGYVDPEYYQCYQLTDKSDVYSFGVVLVELIS 556
F VKVADFGLSR PNDVTHVSTAPQG+PGY+DPEYY CYQLT KSDVYSFGVVL+ELIS
Sbjct: 2 FCVKVADFGLSRDVPNDVTHVSTAPQGSPGYLDPEYYNCYQLTSKSDVYSFGVVLIELIS 181
Query: 557 SLQAVDITRHRNDVNLANMAVNKIQSQELYDLVDPNLGYEKDNSVKRMTTAVAELAFRCL 616
S AVD+ R R+++NL+N+AV KIQ + +LVDP+LG++ D V M +VA LAF+CL
Sbjct: 182 SXPAVDMNRSRDEINLSNLAVRKIQESAVSELVDPSLGFDSDCRVMGMIVSVAGLAFQCL 361
Query: 617 QQQRDLRPSMDEIVEVLRAIKSDEPE 642
Q+++DLRPSM E++ LR I+S + E
Sbjct: 362 QREKDLRPSMYEVLHELRRIESGKDE 439
>TC216865 similar to UP|Q9LPG0 (Q9LPG0) T3F20.24 protein, partial (28%)
Length = 1358
Score = 193 bits (490), Expect = 2e-49
Identities = 117/332 (35%), Positives = 184/332 (55%), Gaps = 11/332 (3%)
Frame = +1
Query: 364 SKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNEVEILARLRHKNLVTLYGC 423
+ ++GEGGFG VYKG DG ++AVK+ + + +F+NE+ +++ L+H +LV LYGC
Sbjct: 4 ANKIGEGGFGPVYKGCFSDGTLIAVKQLSSKSRQGNREFLNEIGMISALQHPHLVKLYGC 183
Query: 424 TSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIALETAEALAYLHASD--- 480
+ +LLLVYEY+ N ++A L G L W+ R I + A LAYLH
Sbjct: 184 CVEGD-QLLLVYEYMENNSLARALFGAEEHQIKLDWTTRYKICVGIARGLAYLHEESRLK 360
Query: 481 VMHRDVKSNNILLDEKFHVKVADFGLSRLFPNDVTHVSTAPQGTPGYVDPEYYQCYQLTD 540
++HRD+K+ N+LLD+ + K++DFGL++L D TH+ST GT GY+ PEY LTD
Sbjct: 361 IVHRDIKATNVLLDQDLNPKISDFGLAKLDEEDNTHISTRIAGTFGYMAPEYAMHGYLTD 540
Query: 541 KSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKIQSQELYDLVDPNLGYEKDNS 600
K+DVYSFG+V +E+I+ + ++ A + ++ DLVD LG E +
Sbjct: 541 KADVYSFGIVALEIINGRSNTIHRQKEESFSVLEWAHLLREKGDIMDLVDRRLGLEFN-- 714
Query: 601 VKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVL--RAIKSDEPETQESKVLD------VV 652
K + ++A C LRP+M +V +L + + +E + ++VLD +
Sbjct: 715 -KEEALVMIKVALLCTNVTAALRPTMSSVVSMLEGKIVVDEEFSGETTEVLDEKKMEKMR 891
Query: 653 VRTDELVLLKKGPYPTSPDSVAEKWVSGSSTS 684
+ EL K+ P+ S SVA+ + G +S
Sbjct: 892 LYYQELSNSKEEPWTASSTSVADLYPVGLDSS 987
>TC217441 homologue to GB|AAK68074.1|14573459|AF384970 somatic embryogenesis
receptor-like kinase 3 {Arabidopsis thaliana;} , partial
(54%)
Length = 1008
Score = 192 bits (487), Expect = 5e-49
Identities = 123/293 (41%), Positives = 173/293 (58%), Gaps = 8/293 (2%)
Frame = +2
Query: 349 FTYEELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRV-AQFMNEVE 407
F+ EL+ AT+NF LG GGFG VYKG L DG +VAVKR E + QF EVE
Sbjct: 119 FSLRELQVATDNFSNKHILGRGGFGKVYKGRLADGSLVAVKRLKEERTQGG*LQFQTEVE 298
Query: 408 ILARLRHKNLVTLYG-CTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIA 466
+++ H+NL+ L G C + R LLVY Y++NG+VA L + S L W R IA
Sbjct: 299 MISMAVHRNLLRLRGFCMTPTER--LLVYPYMANGSVASCLRERQESQPPLGWPERKRIA 472
Query: 467 LETAEALAYLHAS---DVMHRDVKSNNILLDEKFHVKVADFGLSRLFPNDVTHVSTAPQG 523
L +A LAYLH ++HRDVK+ NILLDE+F V DFGL++L THV+TA +G
Sbjct: 473 LGSARGLAYLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFGLAKLMDYKDTHVTTAVRG 652
Query: 524 TPGYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRN--DVNLANMAVNKIQ 581
T G++ PEY + ++K+DV+ +GV+L+ELI+ +A D+ R N DV L + ++
Sbjct: 653 TIGHIAPEYLSTGKSSEKTDVFGYGVMLLELITGQRAFDLARLANDDDVMLLDWVKGLLK 832
Query: 582 SQELYDLVDPNL-GYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVL 633
++L LVD +L G D V+++ ++A C Q RP M E+V +L
Sbjct: 833 DRKLETLVDADLQGSYNDEEVEQL----IQVALLCTQGSPMERPKMSEVVRML 979
>TC232657 weakly similar to UP|O22580 (O22580) Receptor-like serine/threonine
kinase, partial (35%)
Length = 1208
Score = 191 bits (486), Expect = 7e-49
Identities = 102/209 (48%), Positives = 140/209 (66%), Gaps = 3/209 (1%)
Frame = +3
Query: 351 YEELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNEVEILA 410
YE LE+ATN F+ + +LG+GG G+VYKG + DG VA+KR + + F EV +++
Sbjct: 180 YEVLEKATNYFNEANKLGQGGSGSVYKGVMPDGNTVAIKRLSYNTTQWAEHFFTEVNLIS 359
Query: 411 RLRHKNLVTLYGCTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIALETA 470
+ HKNLV L GC S E LLVYEY+ N ++ DH R+S L W +R I L A
Sbjct: 360 GIHHKNLVKLLGC-SITGPESLLVYEYVPNQSLHDHFSVRRTSQPLT-WEMRQKIILGIA 533
Query: 471 EALAYLHASD---VMHRDVKSNNILLDEKFHVKVADFGLSRLFPNDVTHVSTAPQGTPGY 527
E +AYLH ++HRD+K +NILL+E F K+ADFGL+RLFP D +H+STA GT GY
Sbjct: 534 EGMAYLHEESHVRIIHRDIKLSNILLEEDFTPKIADFGLARLFPEDKSHISTAIAGTLGY 713
Query: 528 VDPEYYQCYQLTDKSDVYSFGVVLVELIS 556
+ PEY +LT+K+DVYSFGV+++E++S
Sbjct: 714 MAPEYIVRGKLTEKADVYSFGVLVIEIVS 800
>TC208867 similar to UP|Q9LXT2 (Q9LXT2) Serine/threonine-specific protein
kinase-like protein, partial (72%)
Length = 1118
Score = 191 bits (486), Expect = 7e-49
Identities = 123/279 (44%), Positives = 172/279 (61%), Gaps = 9/279 (3%)
Frame = +1
Query: 364 SKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNEVEILARLRHKNLVTLYGC 423
S +G GGFG VY+G L DGR VA+K ++ + +F EVE+L+RL L+ L G
Sbjct: 7 SNVIGHGGFGLVYRGVLNDGRKVAIKFMDQAGKQGEEEFKVEVELLSRLHSPYLLALLGY 186
Query: 424 TSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLP----WSVRLDIALETAEALAYLH-- 477
S S LLVYE+++NG + +HL+ S+S + P W RL IALE A+ L YLH
Sbjct: 187 CSD-SNHKLLVYEFMANGGLQEHLY-PVSNSIITPVKLDWETRLRIALEAAKGLEYLHEH 360
Query: 478 -ASDVMHRDVKSNNILLDEKFHVKVADFGLSRLFPNDVT-HVSTAPQGTPGYVDPEYYQC 535
+ V+HRD KS+NILLD+KFH KV+DFGL++L P+ HVST GT GYV PEY
Sbjct: 361 VSPPVIHRDFKSSNILLDKKFHAKVSDFGLAKLGPDRAGGHVSTRVLGTQGYVAPEYALT 540
Query: 536 YQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKIQSQE-LYDLVDPNLG 594
LT KSDVYS+GVVL+EL++ VD+ R + L + A+ + +E + ++DP+L
Sbjct: 541 GHLTTKSDVYSYGVVLLELLTGRVPVDMKRPPGEGVLVSWALPLLTDREKVVKIMDPSL- 717
Query: 595 YEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVL 633
E S+K + VA +A C+Q + D RP M ++V+ L
Sbjct: 718 -EGQYSMKEV-VQVAAIAAMCVQPEADYRPLMADVVQSL 828
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.134 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,811,625
Number of Sequences: 63676
Number of extensions: 559314
Number of successful extensions: 5583
Number of sequences better than 10.0: 1020
Number of HSP's better than 10.0 without gapping: 4513
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4642
length of query: 687
length of database: 12,639,632
effective HSP length: 104
effective length of query: 583
effective length of database: 6,017,328
effective search space: 3508102224
effective search space used: 3508102224
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC147875.4


