

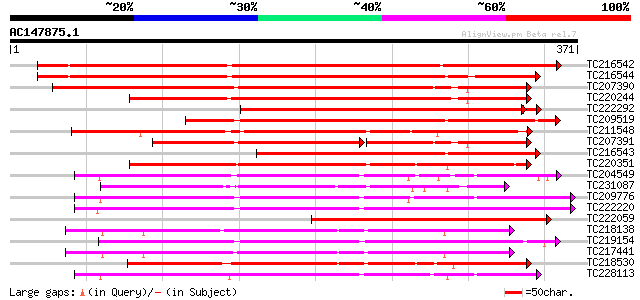
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147875.1 + phase: 0
(371 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC216542 weakly similar to UP|Q9LL55 (Q9LL55) Receptor-like kina... 355 2e-98
TC216544 weakly similar to UP|Q9LL55 (Q9LL55) Receptor-like kina... 332 2e-91
TC207390 weakly similar to UP|Q9ATQ5 (Q9ATQ5) LRK33, partial (34%) 307 6e-84
TC220244 weakly similar to UP|Q9LL58 (Q9LL58) Receptor-like kina... 270 8e-73
TC222292 similar to UP|Q9FID5 (Q9FID5) Receptor protein kinase-l... 255 2e-68
TC209519 similar to UP|Q84K89 (Q84K89) Receptor kinase LRK10 (Re... 241 3e-64
TC211548 weakly similar to UP|Q7XIP7 (Q7XIP7) Receptor-like kina... 240 7e-64
TC207391 similar to UP|Q9SWU8 (Q9SWU8) Receptor-like kinase (Fra... 160 1e-62
TC216543 weakly similar to UP|Q84V32 (Q84V32) Receptor kinase LR... 209 2e-54
TC220351 similar to UP|Q961E7 (Q961E7) GH28523p (CG1830-PB) , pa... 207 6e-54
TC204549 similar to UP|Q9M0X5 (Q9M0X5) Receptor protein kinase-l... 201 4e-52
TC231087 similar to UP|Q39203 (Q39203) Receptor-like protein kin... 201 6e-52
TC209776 similar to UP|O65470 (O65470) Serine/threonine kinase-l... 200 7e-52
TC222220 weakly similar to UP|Q9ZP16 (Q9ZP16) Receptor-like prot... 200 1e-51
TC222059 similar to GB|CAD39676.1|21741394|OSJN00121 OSJNBb0051N... 187 9e-48
TC218138 homologue to UP|Q8GRK2 (Q8GRK2) Somatic embryogenesis r... 182 2e-46
TC219154 similar to PIR|T05754|T05754 S-receptor kinase M4I22.1... 181 6e-46
TC217441 homologue to GB|AAK68074.1|14573459|AF384970 somatic em... 180 8e-46
TC218530 similar to UP|Q9SGU0 (Q9SGU0) T6H22.8.1 protein, partia... 179 1e-45
TC228113 similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kin... 179 1e-45
>TC216542 weakly similar to UP|Q9LL55 (Q9LL55) Receptor-like kinase, partial
(38%)
Length = 1435
Score = 355 bits (911), Expect = 2e-98
Identities = 172/343 (50%), Positives = 240/343 (69%)
Frame = +2
Query: 19 RERTELVDNNVEVFMRSHNLSMPRRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQ 78
R+R + N+E ++ +NL MP YSY E+K + F+EKLG GGYG+V+K L++
Sbjct: 305 RKRHLSIYENIENYLEQNNL-MPIGYSYKEIKKMARGFKEKLGGGGYGIVFKGKLHSGPS 481
Query: 79 VAVKVISETKGNGEEFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFI 138
VA+K++ + KGNG++FI+E+A+I R H N+V L+GYC E +KRAL+YEFMP GSLDKFI
Sbjct: 482 VAIKMLHKAKGNGQDFISEIATIGRIHHQNVVQLIGYCVEGSKRALVYEFMPNGSLDKFI 661
Query: 139 YKSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKIS 198
+ + ++ ++ IAIG+A+G+ YLH GC +ILH DIKP NILLDE F PK+S
Sbjct: 662 FTKD---GNIHLTYDEIYNIAIGVARGIAYLHHGCEMQILHFDIKPHNILLDETFTPKVS 832
Query: 199 DFGLAKICQRKDSIVSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRK 258
DFGLAK+ +SIV++ ARGTIGYMAPE+F + GG+S+++DVYS+GML++EM RK
Sbjct: 833 DFGLAKLYPIDNSIVTMTAARGTIGYMAPELFYKNIGGISHKADVYSFGMLLMEMASKRK 1012
Query: 259 NYDTGGSCTSEMYFPDWIYKDLEQGNTLLNCSTISEEENDMIRKITLVSLWCIQTKPSDR 318
N + +S++YFP WIY L + T + ++EEEN + +K+ +VSLWCIQ KP+DR
Sbjct: 1013NLNPHADHSSQLYFPFWIYNQLGK-ETDIEMEGVTEEENKIAKKMIIVSLWCIQLKPTDR 1189
Query: 319 PPMNKVIEMLQGPLSSVSYPPKPVLYSPERPELQVSDMSSSDL 361
P MNKV+EML+G + S+ PPKP LY E E S SS +
Sbjct: 1190PSMNKVVEMLEGDIESLEIPPKPSLYPHETMENDQSIYSSQTM 1318
>TC216544 weakly similar to UP|Q9LL55 (Q9LL55) Receptor-like kinase, partial
(36%)
Length = 1229
Score = 332 bits (850), Expect = 2e-91
Identities = 163/329 (49%), Positives = 228/329 (68%)
Frame = +2
Query: 19 RERTELVDNNVEVFMRSHNLSMPRRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQ 78
R+R + ++E ++ +NL MP RYSY EVK + F++KLG+GGYG V+K L +
Sbjct: 236 RKRHLSMFESIENYLEQNNL-MPIRYSYKEVKKMAGGFKDKLGEGGYGSVFKGKLGSGSC 412
Query: 79 VAVKVISETKGNGEEFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFI 138
VA+K++ ++KGNG++FI+EVA+I RT H NIV L+G+C +KRAL+YEFMP GSLDKFI
Sbjct: 413 VAIKMLGKSKGNGQDFISEVATIGRTYHQNIVQLIGFCVHGSKRALVYEFMPNGSLDKFI 592
Query: 139 YKSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKIS 198
+ + ++ ++ I+I +A+G+ YLH GC +ILH DIKP NILLDE+F PK+S
Sbjct: 593 FSKD---ESIHLSYDRIYNISIEVARGIAYLHYGCEMQILHFDIKPHNILLDENFTPKVS 763
Query: 199 DFGLAKICQRKDSIVSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRK 258
DFGLAK+ +SIV ARGTIGYMAPE+F GG+S+++DVYSYGML++EM RK
Sbjct: 764 DFGLAKLYPIDNSIVPRTAARGTIGYMAPELFYNNIGGISHKADVYSYGMLLMEMASKRK 943
Query: 259 NYDTGGSCTSEMYFPDWIYKDLEQGNTLLNCSTISEEENDMIRKITLVSLWCIQTKPSDR 318
N + +S+++FP WIY + + ++EEE MI +V+LWCIQ KP+DR
Sbjct: 944 NLNPHAERSSQLFFPFWIYNHIGDEED-IEMEDVTEEEKKMI----IVALWCIQLKPNDR 1108
Query: 319 PPMNKVIEMLQGPLSSVSYPPKPVLYSPE 347
P MNKV+EML+G + ++ PPKP LY E
Sbjct: 1109PSMNKVVEMLEGDIENLEIPPKPTLYPSE 1195
>TC207390 weakly similar to UP|Q9ATQ5 (Q9ATQ5) LRK33, partial (34%)
Length = 1342
Score = 307 bits (786), Expect = 6e-84
Identities = 150/316 (47%), Positives = 216/316 (67%), Gaps = 3/316 (0%)
Frame = +3
Query: 29 VEVFMRSHNLSMPRRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVISETK 88
V F+ + P R++YA++K IT F+EKLG+G +G V++ L N VAVK+++ T+
Sbjct: 168 VAKFLEDYRAEKPARFTYADLKRITGGFKEKLGEGAHGAVFRGKLSNEILVAVKILNNTE 347
Query: 89 GNGEEFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAIC 148
G G+EFINEV + + H+N+V LLG+C E RAL+Y P GSL +FI + +
Sbjct: 348 GEGKEFINEVGIMGKIHHINVVRLLGFCAEGFHRALVYNLFPNGSLQRFIVPPDDKDHF- 524
Query: 149 DFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQR 208
W L +IA+GIAKG+EYLH+GC+ I+H DI P N+LLD++F PKISDFGLAK+C +
Sbjct: 525 -LGWEKLQQIALGIAKGIEYLHEGCNQPIIHFDINPHNVLLDDNFTPKISDFGLAKLCSK 701
Query: 209 KDSIVSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTS 268
S+VS+ ARGT+GY+APE+FS+ FG VSY+SD+YSYGML+LEM+GGRKN D +
Sbjct: 702 NPSLVSMTAARGTLGYIAPEVFSKNFGNVSYKSDIYSYGMLLLEMVGGRKNVDMSSAQDF 881
Query: 269 EMYFPDWIYKDLEQGNTLLNCSTISEEEND--MIRKITLVSLWCIQTKPSDRPPMNKVIE 326
+ +PDWI+ +L G+ ++ E+E D + +K+ +V LWCIQ +P +RP + VI+
Sbjct: 882 HVLYPDWIH-NLIDGDVHIHV----EDECDIKIAKKLAIVGLWCIQWQPVNRPSIKSVIQ 1046
Query: 327 MLQ-GPLSSVSYPPKP 341
ML+ G S ++ PP P
Sbjct: 1047MLETGGESQLNVPPNP 1094
>TC220244 weakly similar to UP|Q9LL58 (Q9LL58) Receptor-like kinase, partial
(32%)
Length = 1112
Score = 270 bits (690), Expect = 8e-73
Identities = 129/265 (48%), Positives = 183/265 (68%), Gaps = 2/265 (0%)
Frame = +1
Query: 79 VAVKVISETKGNGEEFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFI 138
VAVK++++T G+G++FINEV ++ + H+N+V LLG+C + RAL+Y+F P GSL +F+
Sbjct: 40 VAVKILNDTVGDGKDFINEVGTMGKIHHVNVVRLLGFCADGFHRALVYDFFPNGSLQRFL 219
Query: 139 YKSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKIS 198
+ N W L +IA+G+AKG+EYLH GC RILH DI P NIL+D+ F PKI+
Sbjct: 220 APPD--NKDVFLGWEKLQQIALGVAKGVEYLHLGCDQRILHFDINPHNILIDDHFVPKIT 393
Query: 199 DFGLAKICQRKDSIVSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRK 258
DFGLAK+C + S VSI ARGT+GY+APE+FSR FG VSY+SD+YSYGML+LEM+GGRK
Sbjct: 394 DFGLAKLCPKNQSTVSITAARGTLGYIAPEVFSRNFGNVSYKSDIYSYGMLLLEMVGGRK 573
Query: 259 NYDTGGSCTSEMYFPDWIYKDLEQGNTLLNCSTISEEEND--MIRKITLVSLWCIQTKPS 316
N + + ++ +P+WI+ L+ + + E+E D + +K+ +V LWCI+ P
Sbjct: 574 NTNMSAEESFQVLYPEWIHNLLKSRDVQVTI----EDEGDVRIAKKLAIVGLWCIEWNPI 741
Query: 317 DRPPMNKVIEMLQGPLSSVSYPPKP 341
DRP M VI+ML+G + PP P
Sbjct: 742 DRPSMKTVIQMLEGDGDKLIAPPTP 816
>TC222292 similar to UP|Q9FID5 (Q9FID5) Receptor protein kinase-like protein,
partial (16%)
Length = 732
Score = 255 bits (651), Expect(2) = 2e-68
Identities = 113/187 (60%), Positives = 152/187 (80%)
Frame = +2
Query: 152 WNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDS 211
W+ ++IAIGIA+GLEYLH+GC++RILH DIKP NILLDE+FCPKISDFGLAK+C RK S
Sbjct: 8 WDNFWQIAIGIARGLEYLHRGCNTRILHFDIKPHNILLDENFCPKISDFGLAKLCPRKGS 187
Query: 212 IVSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMY 271
I+S+ RGTIGY+APE+++R FGGVS++SDVY YGM++LEM+GGR N + S TSE+Y
Sbjct: 188 IISMSDPRGTIGYVAPEVWNRNFGGVSHKSDVYRYGMMLLEMVGGRNNINAEASHTSEIY 367
Query: 272 FPDWIYKDLEQGNTLLNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGP 331
FPDWIYK LEQG L ++ EEN++++++ +V LWC+QT P DRP M +V++ML+G
Sbjct: 368 FPDWIYKRLEQGGDLRPNGVMAPEENEIVKRMPVVGLWCVQTFPKDRPAMTRVVDMLEGK 547
Query: 332 LSSVSYP 338
++S+ P
Sbjct: 548 MNSLDIP 568
Score = 22.3 bits (46), Expect(2) = 2e-68
Identities = 9/12 (75%), Positives = 9/12 (75%)
Frame = +1
Query: 337 YPPKPVLYSPER 348
YP KPVL SP R
Sbjct: 562 YPQKPVLSSPTR 597
>TC209519 similar to UP|Q84K89 (Q84K89) Receptor kinase LRK10 (Receptor
kinase LRK14), partial (19%)
Length = 866
Score = 241 bits (616), Expect = 3e-64
Identities = 117/245 (47%), Positives = 164/245 (66%)
Frame = +2
Query: 116 CYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSS 175
C E K AL+YEFMP GSLDK+I+ E ++ ++ + I +GIA+G+ YLHQ C
Sbjct: 2 CAEGEKHALVYEFMPNGSLDKYIFSKEESVSL---SYDKTYEICLGIARGIAYLHQDCDV 172
Query: 176 RILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARGTIGYMAPEIFSRAFG 235
+ILH DIKP NILLD++F PK+SDFGLAK+ KD + + G RGT GYMAPE+F + G
Sbjct: 173 QILHFDIKPHNILLDDNFIPKVSDFGLAKLYPIKDKSIILTGLRGTFGYMAPELFYKNIG 352
Query: 236 GVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMYFPDWIYKDLEQGNTLLNCSTISEE 295
GVSY++DVYS+GML++EM R+N + +S+ +FP WIY + ++ +SEE
Sbjct: 353 GVSYKADVYSFGMLLMEMGSRRRNSNPHTEHSSQHFFPFWIYDHFMEEKD-IHMEEVSEE 529
Query: 296 ENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPLSSVSYPPKPVLYSPERPELQVSD 355
+ +++K+ +VSLWCIQ KP+DRP M KV+EML+G + ++ PPKPV Y P +
Sbjct: 530 DKILVKKMFIVSLWCIQLKPNDRPSMKKVVEMLEGKVENIDMPPKPVFY-PHETTIDSDQ 706
Query: 356 MSSSD 360
S SD
Sbjct: 707 ASWSD 721
>TC211548 weakly similar to UP|Q7XIP7 (Q7XIP7) Receptor-like kinase
TAK33-like protein, partial (58%)
Length = 1194
Score = 240 bits (613), Expect = 7e-64
Identities = 126/306 (41%), Positives = 197/306 (64%), Gaps = 4/306 (1%)
Frame = +2
Query: 41 PRRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVI--SETKGNGEEFINEV 98
P R++ ++++ T+ + LG GG+GVVYK S N VAVKV+ S K E+F+ EV
Sbjct: 5 PIRFTDQQLRIATDNYSFLLGSGGFGVVYKGSFSNGTIVAVKVLRGSSDKRIDEQFMAEV 184
Query: 99 ASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRI 158
+I + H N+V L G+C+E + RAL+YE+M G+L+K+++ N F+ L I
Sbjct: 185 GTIGKVHHFNLVRLYGFCFERHLRALVYEYMVNGALEKYLFHE---NTTLSFE--KLHEI 349
Query: 159 AIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGA 218
A+G A+G+ YLH+ C RI+H DIKP NILLD +FCPK++DFGLAK+C R ++ +S+ G
Sbjct: 350 AVGTARGIAYLHEECQQRIIHYDIKPGNILLDRNFCPKVADFGLAKLCNRDNTHISMTGG 529
Query: 219 RGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMYFPDWIYK 278
RGT GY APE++ V+++ DVYS+GML+ E+IG R+N++ S+++FP W+++
Sbjct: 530 RGTPGYAAPELWLPF--PVTHKCDVYSFGMLLFEIIGRRRNHNI-NLPESQVWFPMWVWE 700
Query: 279 --DLEQGNTLLNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPLSSVS 336
D E L++ I ++ ++ +I V+L C+Q KP RP M+ V++ML+G +
Sbjct: 701 RFDAENVEDLISACGIEDQNREIAERIVKVALSCVQYKPEARPIMSVVVKMLEGSVE--- 871
Query: 337 YPPKPV 342
PKP+
Sbjct: 872 -VPKPL 886
>TC207391 similar to UP|Q9SWU8 (Q9SWU8) Receptor-like kinase (Fragment),
partial (29%)
Length = 963
Score = 160 bits (406), Expect(2) = 1e-62
Identities = 76/139 (54%), Positives = 97/139 (69%)
Frame = +1
Query: 94 FINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWN 153
FINEV + + H+N+V LLGYC E RAL+Y F P GSL FI+ + W
Sbjct: 19 FINEVEIMGKIHHINVVRLLGYCAEGIHRALVYNFFPNGSLQSFIFPPDDKQNF--LGWE 192
Query: 154 TLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIV 213
L IA+GIAKG+ YLHQGC+ I+H DI P N+LLD++F PKISDFGLAK+C + S+V
Sbjct: 193 KLQNIALGIAKGIGYLHQGCNHPIIHFDINPHNVLLDDNFTPKISDFGLAKLCSKNPSLV 372
Query: 214 SILGARGTIGYMAPEIFSR 232
S+ ARGT+GY+APE+FSR
Sbjct: 373 SMTAARGTLGYIAPEVFSR 429
Score = 97.4 bits (241), Expect(2) = 1e-62
Identities = 50/111 (45%), Positives = 72/111 (64%), Gaps = 3/111 (2%)
Frame = +2
Query: 234 FGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMYFPDWIYKDLEQGNTLLNCSTIS 293
FG VSY+SD+YSYGML+LEM+GGRKN DT + +PDW++ DL G+ ++
Sbjct: 434 FGNVSYKSDIYSYGMLLLEMVGGRKNVDTSSPEDFHVLYPDWMH-DLVHGDVHIHV---- 598
Query: 294 EEEND--MIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPLSS-VSYPPKP 341
E+E D + RK+ +V LWCIQ +P +RP + VI+ML+ ++ PP P
Sbjct: 599 EDEGDVKIARKLAIVGLWCIQWQPLNRPSIKSVIQMLESKEEDLLTVPPNP 751
>TC216543 weakly similar to UP|Q84V32 (Q84V32) Receptor kinase LRK45, partial
(23%)
Length = 603
Score = 209 bits (532), Expect = 2e-54
Identities = 96/186 (51%), Positives = 131/186 (69%)
Frame = +2
Query: 162 IAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARGT 221
+A+G+ YLH GC +ILH DIKP N LDE+F PK+SDFGLAK+ +SIV ARGT
Sbjct: 2 VARGIAYLHYGCEMQILHFDIKPHNXXLDENFTPKVSDFGLAKLYPIDNSIVPRTAARGT 181
Query: 222 IGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMYFPDWIYKDLE 281
IGYMAPE+F GG+S+++DVYSYGML++EM RKN + +S+++FP WIY +
Sbjct: 182 IGYMAPELFYNNIGGISHKADVYSYGMLLMEMASKRKNLNPHAERSSQLFFPFWIYNHIG 361
Query: 282 QGNTLLNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPLSSVSYPPKP 341
+ ++EEE M++K+ +V+LWCIQ KP+DRP MNKV+EML+G + ++ PPKP
Sbjct: 362 DEED-IEMEDVTEEEKKMVKKMIIVALWCIQLKPNDRPSMNKVVEMLEGDIENLEIPPKP 538
Query: 342 VLYSPE 347
LY E
Sbjct: 539 TLYPSE 556
>TC220351 similar to UP|Q961E7 (Q961E7) GH28523p (CG1830-PB) , partial (11%)
Length = 1064
Score = 207 bits (527), Expect = 6e-54
Identities = 111/267 (41%), Positives = 169/267 (62%), Gaps = 4/267 (1%)
Frame = +1
Query: 79 VAVKVISETKGNGEEFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFI 138
VAVK + + ++F EVA+IS T H+N+V L+G+C E R L+YEFM GSLD F+
Sbjct: 1 VAVKQLEGIEQGEKQFRMEVATISSTHHLNLVRLIGFCSEGRHRLLVYEFMKNGSLDDFL 180
Query: 139 YKSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKIS 198
+ +E + +W F IA+G A+G+ YLH+ C I+H DIKP+NILLDE++ K+S
Sbjct: 181 FLTEQHSGKL-LNWEYRFNIALGTARGITYLHEECRDCIVHCDIKPENILLDENYVAKVS 357
Query: 199 DFGLAKICQRKD-SIVSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGR 257
DFGLAK+ KD ++ RGT GY+APE + ++ +SDVY YGM++LE++ GR
Sbjct: 358 DFGLAKLINPKDHRHRTLTSVRGTRGYLAPEWLANL--PITSKSDVYGYGMVLLEIVSGR 531
Query: 258 KNYDTGGSCTSEMYFPDWIYKDLEQGNT--LLNCSTISEE-ENDMIRKITLVSLWCIQTK 314
+N+D T+ F W Y++ E+GN +L+ ++E + + +R+ S WCIQ +
Sbjct: 532 RNFDVSEE-TNRKKFSIWAYEEFEKGNISGILDKRLANQEVDMEQVRRAIQASFWCIQEQ 708
Query: 315 PSDRPPMNKVIEMLQGPLSSVSYPPKP 341
PS RP M++V++ML+G ++ PP P
Sbjct: 709 PSHRPTMSRVLQMLEG-VTEPERPPAP 786
>TC204549 similar to UP|Q9M0X5 (Q9M0X5) Receptor protein kinase-like protein,
partial (54%)
Length = 2032
Score = 201 bits (511), Expect = 4e-52
Identities = 125/343 (36%), Positives = 197/343 (56%), Gaps = 24/343 (6%)
Frame = +3
Query: 43 RYSYAEVKMITNYFR--EKLGQGGYGVVYKASLYNSRQVAVKVISETKGNG-EEFINEVA 99
++ ++ ++ TN F KLG+GG+G VYK +L + + VAVK +S++ G G EEF NEV
Sbjct: 789 QFDFSTIEAATNKFSADNKLGEGGFGEVYKGTLSSGQVVAVKRLSKSSGQGGEEFKNEVV 968
Query: 100 SISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIA 159
+++ H N+V LLG+C + ++ L+YE++P SLD ++ E + DW ++I
Sbjct: 969 VVAKLQHRNLVRLLGFCLQGEEKILVYEYVPNKSLDYILFDPEKQR---ELDWGRRYKII 1139
Query: 160 IGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGAR 219
GIA+G++YLH+ RI+H D+K NILLD D PKISDFG+A+I + +
Sbjct: 1140 GGIARGIQYLHEDSRLRIIHRDLKASNILLDGDMNPKISDFGMARIFGVDQTQGNTSRIV 1319
Query: 220 GTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKN---YDTGGSCTSEMYFPDW- 275
GT GYMAPE G S +SDVYS+G+L++E++ G+KN Y T G+ ++ W
Sbjct: 1320 GTYGYMAPEY--AMHGEFSVKSDVYSFGVLLMEILSGKKNSSFYQTDGA--EDLLSYAWQ 1487
Query: 276 IYKD---LEQGNTLLNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPL 332
++KD LE + +L S +N++IR I + L C+Q P+DRP M ++ ML
Sbjct: 1488 LWKDGTPLELMDPILR---ESYNQNEVIRSIH-IGLLCVQEDPADRPTMATIVLMLDSNT 1655
Query: 333 SSVSYPPKPVLY-----SPERPE---------LQVSDMSSSDL 361
++ P +P + P P+ + V+DMS S++
Sbjct: 1656 VTLPTPTQPAFFVHSGTDPNMPKELPFDQSIPMSVNDMSISEM 1784
>TC231087 similar to UP|Q39203 (Q39203) Receptor-like protein kinase
precursor, partial (30%)
Length = 826
Score = 201 bits (510), Expect = 6e-52
Identities = 118/288 (40%), Positives = 169/288 (57%), Gaps = 20/288 (6%)
Frame = +1
Query: 60 LGQGGYGVVYKASLYNSRQVAVKVISETKGNGEEFINEVASISRTSHMNIVSLLGYCYEE 119
+G GG+G V++ L ++ VAVK + G +EF EV++I H+N+V L G+C E
Sbjct: 1 VGHGGFGTVFQGELSDASVVAVKRLERPGGGEKEFRAEVSTIGNIQHVNLVRLRGFCSEN 180
Query: 120 NKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSSRILH 179
+ R L+YE+M G+L+ ++ K E P C W+ FR+A+G AKG+ YLH+ C I+H
Sbjct: 181 SHRLLVYEYMQNGALNVYLRK-EGP---C-LSWDVRFRVAVGTAKGIAYLHEECRCCIIH 345
Query: 180 LDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARGTIGYMAPEIFSRAFGGVSY 239
DIKP+NILLD DF K+SDFGLAK+ R S V ++ RGT GY+APE S ++
Sbjct: 346 CDIKPENILLDGDFTAKVSDFGLAKLIGRDFSRV-LVTMRGTWGYVAPEWISGV--AITT 516
Query: 240 RSDVYSYGMLILEMIGGRKNYDT---------GGSCTSEM----YFPDWIYKDLEQGNT- 285
++DVYSYGM +LE+IGGR+N + GG EM +FP W + + +GN
Sbjct: 517 KADVYSYGMTLLELIGGRRNVEAPLSAGGGGGGGESGDEMGGKWFFPPWAAQRIIEGNVS 696
Query: 286 ------LLNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEM 327
L N I E R++ LV++WCIQ + RP M V++M
Sbjct: 697 DVMDKRLGNAYNIEE-----ARRVALVAVWCIQDDEAMRPTMGMVVKM 825
>TC209776 similar to UP|O65470 (O65470) Serine/threonine kinase-like protein
, partial (47%)
Length = 1305
Score = 200 bits (509), Expect = 7e-52
Identities = 124/335 (37%), Positives = 186/335 (55%), Gaps = 7/335 (2%)
Frame = +2
Query: 43 RYSYAEVKMITNYFRE--KLGQGGYGVVYKASLYNSRQVAVKVISETKGNG-EEFINEVA 99
R+ ++ ++ T F E KLG+GG+G VYK L + ++VAVK +S+ G G EEF NEV
Sbjct: 41 RFDFSTIEAATQKFSEANKLGEGGFGEVYKGLLPSGQEVAVKRLSKISGQGGEEFKNEVE 220
Query: 100 SISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIA 159
+++ H N+V LLG+C E ++ L+YEF+ SLD ++ E ++ DW ++I
Sbjct: 221 IVAKLQHRNLVRLLGFCLEGEEKILVYEFVANKSLDYILFDPEKQKSL---DWTRRYKIV 391
Query: 160 IGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGAR 219
GIA+G++YLH+ +I+H D+K N+LLD D PKISDFG+A+I + +
Sbjct: 392 EGIARGIQYLHEDSRLKIIHRDLKASNVLLDGDMNPKISDFGMARIFGVDQTQANTNRIV 571
Query: 220 GTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKN---YDTGGSCTSEMYFPDW- 275
GT GYM+PE G S +SDVYS+G+LILE+I G++N Y+T ++ W
Sbjct: 572 GTYGYMSPEY--AMHGEYSAKSDVYSFGVLILEIISGKRNSSFYET--DVAEDLLSYAWK 739
Query: 276 IYKDLEQGNTLLNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPLSSV 335
++KD + S N++IR I + L C+Q P DRP M V+ ML ++
Sbjct: 740 LWKDEAPLELMDQSLRESYTRNEVIRCIH-IGLLCVQEDPIDRPTMASVVLMLDSYSVTL 916
Query: 336 SYPPKPVLYSPERPELQVSDMSSSDLYETNSVTVS 370
P +P Y R E + D TNS + S
Sbjct: 917 QVPNQPAFYINSRTEPNMPKGLKIDQSTTNSTSKS 1021
>TC222220 weakly similar to UP|Q9ZP16 (Q9ZP16) Receptor-like protein kinase,
RLK3 precursor, partial (35%)
Length = 1304
Score = 200 bits (508), Expect = 1e-51
Identities = 114/332 (34%), Positives = 189/332 (56%), Gaps = 4/332 (1%)
Frame = +3
Query: 43 RYSYAEVKMITNYF--REKLGQGGYGVVYKASLYNSRQVAVKVISETKGNG-EEFINEVA 99
++ ++ TN F +++G+GG+GVVYK L + R++AVK +S++ G G EF NE+
Sbjct: 186 QFGLVTIEAATNKFSYEKRIGEGGFGVVYKGVLPDGREIAVKKLSKSSGQGANEFKNEIL 365
Query: 100 SISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIA 159
I++ H N+V+LLG+C EE+++ LIYEF+ SLD F++ S + +W+ ++I
Sbjct: 366 LIAKLQHRNLVTLLGFCLEEHEKMLIYEFVSNKSLDYFLFDSHRSKQL---NWSERYKII 536
Query: 160 IGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGAR 219
GIA+G+ YLH+ +++H D+KP N+LLD + PKISDFG+A+I
Sbjct: 537 EGIAQGISYLHEHSRLKVIHRDLKPSNVLLDSNMNPKISDFGMARIVAIDQLQGKTNRIV 716
Query: 220 GTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMYFPDWIYKD 279
GT GYM+PE G S +SDV+S+G+++LE+I ++N + S ++ W
Sbjct: 717 GTYGYMSPEY--AMHGQFSEKSDVFSFGVIVLEIISAKRNTRSVFSDHDDLLSYTWEQWM 890
Query: 280 LEQGNTLLNCSTISEE-ENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPLSSVSYP 338
E + + S +E ++ + K + L C+Q KP DRP + +VI L ++ + P
Sbjct: 891 DEAPLNIFDQSIEAEFCDHSEVVKCIQIGLLCVQEKPDDRPKITQVISYLNSSITELPLP 1070
Query: 339 PKPVLYSPERPELQVSDMSSSDLYETNSVTVS 370
KP+ S ++ V + SS N ++VS
Sbjct: 1071KKPIRQSGIVQKIAVGESSSGSTPSINEMSVS 1166
>TC222059 similar to GB|CAD39676.1|21741394|OSJN00121 OSJNBb0051N19.7 {Oryza
sativa (japonica cultivar-group);} , partial (30%)
Length = 510
Score = 187 bits (474), Expect = 9e-48
Identities = 89/157 (56%), Positives = 116/157 (73%)
Frame = +2
Query: 198 SDFGLAKICQRKDSIVSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGR 257
SDFGLAKIC RK+S++SI GARGT GY+APE+FSR FG VS++SDVYSYGM+ILEM+G R
Sbjct: 2 SDFGLAKICTRKESMISIFGARGTAGYIAPEVFSRNFGTVSHKSDVYSYGMMILEMVGRR 181
Query: 258 KNYDTGGSCTSEMYFPDWIYKDLEQGNTLLNCSTISEEENDMIRKITLVSLWCIQTKPSD 317
KN T +C+SE+YFPDWIY LE L + +E ++ ++RK+T+V LWCIQT PS
Sbjct: 182 KNIKTEVNCSSEIYFPDWIYNRLESNEELGLQNIRNESDDKLVRKMTIVGLWCIQTHPST 361
Query: 318 RPPMNKVIEMLQGPLSSVSYPPKPVLYSPERPELQVS 354
RP ++KV+EML + + PPKP L SP + +S
Sbjct: 362 RPAISKVLEMLGSKVELLQIPPKPFLSSPPTSPVHLS 472
>TC218138 homologue to UP|Q8GRK2 (Q8GRK2) Somatic embryogenesis receptor
kinase 1, partial (60%)
Length = 1444
Score = 182 bits (463), Expect = 2e-46
Identities = 111/301 (36%), Positives = 170/301 (55%), Gaps = 7/301 (2%)
Frame = +1
Query: 37 NLSMPRRYSYAEVKMITNYFREK--LGQGGYGVVYKASLYNSRQVAVKVISE--TKGNGE 92
+L +R+S E+++ T+ F K LG+GG+G VYK L + VAVK + E T G
Sbjct: 100 HLGQLKRFSLRELQVATDSFSNKNILGRGGFGKVYKGRLADGSLVAVKRLKEERTPGGEL 279
Query: 93 EFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDW 152
+F EV IS H N++ L G+C +R L+Y +M GS+ + E P DW
Sbjct: 280 QFQTEVEMISMAVHRNLLRLRGFCMTPTERLLVYPYMANGSVASCL--RERPPYQEPLDW 453
Query: 153 NTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSI 212
T R+A+G A+GL YLH C +I+H D+K NILLDE+F + DFGLAK+ KD+
Sbjct: 454 PTRKRVALGSARGLSYLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFGLAKLMDYKDTH 633
Query: 213 VSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCT-SEMY 271
V+ RGTIG++APE S G S ++DV+ YG+++LE+I G++ +D ++
Sbjct: 634 VT-TAVRGTIGHIAPEYLST--GKSSEKTDVFGYGIMLLELITGQRAFDLARLANDDDVM 804
Query: 272 FPDWIYKDLEQG--NTLLNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQ 329
DW+ L++ L++ + + ++ V+L C Q P DRP M++V+ ML+
Sbjct: 805 LLDWVKGLLKEKKLEMLVDPDLQTNYIETEVEQLIQVALLCTQGSPMDRPKMSEVVRMLE 984
Query: 330 G 330
G
Sbjct: 985 G 987
>TC219154 similar to PIR|T05754|T05754 S-receptor kinase M4I22.110 precursor
- Arabidopsis thaliana {Arabidopsis thaliana;} ,
partial (31%)
Length = 1363
Score = 181 bits (458), Expect = 6e-46
Identities = 116/307 (37%), Positives = 166/307 (53%), Gaps = 5/307 (1%)
Frame = +1
Query: 59 KLGQGGYGVVYKASLYNSRQVAVKVISETKGNG-EEFINEVASISRTSHMNIVSLLGYCY 117
K+G+GG+G VYK L +++AVK +S G G EFI EV I++ H N+V LLG C
Sbjct: 364 KIGEGGFGPVYKGKLVGGQEIAVKRLSSLSGQGITEFITEVKLIAKLQHRNLVKLLGCCI 543
Query: 118 EENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSSRI 177
+ ++ L+YE++ GSL+ FI+ + DW F I +GIA+GL YLHQ RI
Sbjct: 544 KGQEKLLVYEYVVNGSLNSFIFDQIKSKLL---DWPRRFNIILGIARGLLYLHQDSRLRI 714
Query: 178 LHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARGTIGYMAPEIFSRAF-GG 236
+H D+K N+LLDE PKISDFG+A+ + + GT GYMAPE AF G
Sbjct: 715 IHRDLKASNVLLDEKLNPKISDFGMARAFGGDQTEGNTNRVVGTYGYMAPEY---AFDGN 885
Query: 237 VSYRSDVYSYGMLILEMIGGRKNYD-TGGSCTSEMYFPDWIYKDLEQGNTLLNCSTISEE 295
S +SDV+S+G+L+LE++ G KN + T + W + L++
Sbjct: 886 FSIKSDVFSFGILLLEIVCGIKNKSLCHENQTLNLVGYAWALWKEQNALQLIDSGIKDSC 1065
Query: 296 ENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPLSSVSYPPKPVLYSPER--PELQV 353
+ + VSL C+Q P DRP M VI+ML + V PK + P R E +
Sbjct: 1066VIPEVLRCIHVSLLCVQQYPEDRPTMTSVIQMLGSEMDMVE--PKEPGFFPRRILKEGNL 1239
Query: 354 SDMSSSD 360
+M+S+D
Sbjct: 1240KEMTSND 1260
>TC217441 homologue to GB|AAK68074.1|14573459|AF384970 somatic embryogenesis
receptor-like kinase 3 {Arabidopsis thaliana;} , partial
(54%)
Length = 1008
Score = 180 bits (457), Expect = 8e-46
Identities = 108/301 (35%), Positives = 170/301 (55%), Gaps = 7/301 (2%)
Frame = +2
Query: 37 NLSMPRRYSYAEVKMITNYFREK--LGQGGYGVVYKASLYNSRQVAVKVISE--TKGNGE 92
+L +R+S E+++ T+ F K LG+GG+G VYK L + VAVK + E T+G
Sbjct: 98 HLGQLKRFSLRELQVATDNFSNKHILGRGGFGKVYKGRLADGSLVAVKRLKEERTQGG*L 277
Query: 93 EFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDW 152
+F EV IS H N++ L G+C +R L+Y +M GS+ + + + W
Sbjct: 278 QFQTEVEMISMAVHRNLLRLRGFCMTPTERLLVYPYMANGSVASCLRERQESQP--PLGW 451
Query: 153 NTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSI 212
RIA+G A+GL YLH C +I+H D+K NILLDE+F + DFGLAK+ KD+
Sbjct: 452 PERKRIALGSARGLAYLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFGLAKLMDYKDTH 631
Query: 213 VSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCT-SEMY 271
V+ RGTIG++APE S G S ++DV+ YG+++LE+I G++ +D ++
Sbjct: 632 VT-TAVRGTIGHIAPEYLST--GKSSEKTDVFGYGVMLLELITGQRAFDLARLANDDDVM 802
Query: 272 FPDWIYKDLEQG--NTLLNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQ 329
DW+ L+ TL++ ++ + ++ V+L C Q P +RP M++V+ ML+
Sbjct: 803 LLDWVKGLLKDRKLETLVDADLQGSYNDEEVEQLIQVALLCTQGSPMERPKMSEVVRMLE 982
Query: 330 G 330
G
Sbjct: 983 G 985
>TC218530 similar to UP|Q9SGU0 (Q9SGU0) T6H22.8.1 protein, partial (25%)
Length = 1190
Score = 179 bits (455), Expect = 1e-45
Identities = 99/267 (37%), Positives = 162/267 (60%), Gaps = 3/267 (1%)
Frame = +3
Query: 78 QVAVKVISETKGNGE-EFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDK 136
++AVK +S G+ +FI E+A+IS H N+V L G C E +KR L+YE++ SLD+
Sbjct: 3 EIAVKQLSVGSHQGKSQFITEIATISAVQHRNLVKLYGCCIEGSKRLLVYEYLENKSLDQ 182
Query: 137 FIYKSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPK 196
++ +W+T + I +G+A+GL YLH+ RI+H D+K NILLD + PK
Sbjct: 183 ALF-----GKCLTLNWSTRYDICLGVARGLTYLHEESRLRIVHRDVKASNILLDYELIPK 347
Query: 197 ISDFGLAKICQRKDSIVSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGG 256
ISDFGLAK+ K + +S G GTIGY+APE R G ++ ++DV+S+G++ LE++ G
Sbjct: 348 ISDFGLAKLYDDKKTHIS-TGVAGTIGYLAPEYAMR--GHLTEKADVFSFGVVALELVSG 518
Query: 257 RKNYDTGGSCTSEMYFPDWIYKDLEQGNTLLNC--STISEEENDMIRKITLVSLWCIQTK 314
R N D+ ++Y +W ++ L + N +++ +SE + ++++ ++L C QT
Sbjct: 519 RPNSDSSLE-GEKVYLLEWAWQ-LHEKNCIIDLVDDRLSEFNEEEVKRVVGIALLCTQTS 692
Query: 315 PSDRPPMNKVIEMLQGPLSSVSYPPKP 341
P+ RP M++V+ ML G + + KP
Sbjct: 693 PTLRPSMSRVVAMLSGDIEVSTVTSKP 773
>TC228113 similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kinase homolog
RK20-1, partial (53%)
Length = 1492
Score = 179 bits (455), Expect = 1e-45
Identities = 113/345 (32%), Positives = 176/345 (50%), Gaps = 39/345 (11%)
Frame = +1
Query: 43 RYSYAEVKMITNYFRE--KLGQGGYGVVYKASLYNSRQVAVKVISETKGNGE-EFINEVA 99
++++ +++ T F + KLGQGG+G VY+ L + + +AVK +S G+ EF NEV
Sbjct: 115 QFNFDTIRVATEDFSDSNKLGQGGFGAVYRGRLSDGQMIAVKRLSRESSQGDTEFKNEVL 294
Query: 100 SISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSE----------------- 142
+++ H N+V LLG+C E +R LIYE++P SLD FI+
Sbjct: 295 LVAKLQHRNLVRLLGFCLEGKERLLIYEYVPNKSLDYFIFGRS**VHN*TVALYFTSGSG 474
Query: 143 ----------------FPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQN 186
P +W ++I G+A+GL YLH+ RI+H D+K N
Sbjct: 475 QRLNIHIPLKLMAVYADPTKKAQLNWEMRYKIITGVARGLLYLHEDSHLRIIHRDLKASN 654
Query: 187 ILLDEDFCPKISDFGLAKICQRKDSIVSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSY 246
ILL+E+ PKI+DFG+A++ + + GT GYMAPE G S +SDV+S+
Sbjct: 655 ILLNEEMNPKIADFGMARLVLMDQTQANTNRIVGTYGYMAPEY--AMHGQFSMKSDVFSF 828
Query: 247 GMLILEMIGGRKNYD-TGGSCTSEMYFPDWIYKDLEQGNT--LLNCSTISEEENDMIRKI 303
G+L+LE+I G KN G ++ W ++ +G +++ S + N+M+R I
Sbjct: 829 GVLVLEIISGHKNSGIRHGENVEDLLSFAW--RNWREGTAVKIVDPSLNNNSRNEMLRCI 1002
Query: 304 TLVSLWCIQTKPSDRPPMNKVIEMLQGPLSSVSYPPKPVLYSPER 348
+ L C+Q +DRP M ++ ML S+ P KP Y R
Sbjct: 1003H-IGLLCVQENLADRPTMTTIMLMLNSYSLSLPIPSKPAFYVSSR 1134
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.136 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,268,255
Number of Sequences: 63676
Number of extensions: 242498
Number of successful extensions: 3061
Number of sequences better than 10.0: 958
Number of HSP's better than 10.0 without gapping: 2153
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2182
length of query: 371
length of database: 12,639,632
effective HSP length: 99
effective length of query: 272
effective length of database: 6,335,708
effective search space: 1723312576
effective search space used: 1723312576
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC147875.1


