

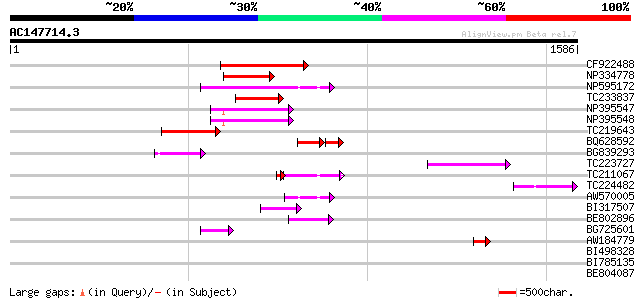
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147714.3 - phase: 0 /pseudo
(1586 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CF922488 329 7e-90
NP334778 reverse transcriptase [Glycine max] 221 3e-57
NP595172 polyprotein [Glycine max] 215 1e-55
TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 206 9e-53
NP395547 reverse transcriptase [Glycine max] 183 6e-46
NP395548 reverse transcriptase [Glycine max] 174 2e-43
TC219643 weakly similar to UP|Q6WAY7 (Q6WAY7) Gag/pol polyprotei... 166 8e-41
BQ628592 100 3e-38
BG839293 112 1e-24
TC223727 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 107 3e-23
TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%) 53 9e-12
TC224482 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 70 1e-11
AW570005 65 2e-10
BI317507 65 2e-10
BE802896 59 1e-08
BG725601 similar to PIR|H86337|H86 protein F5M15.26 [imported] -... 59 2e-08
AW184779 47 5e-05
BI498328 39 0.014
BI785135 39 0.024
BE804087 37 0.069
>CF922488
Length = 741
Score = 329 bits (843), Expect = 7e-90
Identities = 165/246 (67%), Positives = 192/246 (77%)
Frame = +3
Query: 589 VPKKDGKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSP 648
V K+DGKV MCVD+RDLN ASPKD FPLPHI+VLVDNT FSFMDGFSGYNQIK++P
Sbjct: 3 VLKEDGKV*MCVDYRDLN*ASPKDKFPLPHINVLVDNTTSFSQFSFMDGFSGYNQIKIAP 182
Query: 649 EDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKD 708
ED EKT+FIT WGTFCYK M FGL N GATYQR M LF DM+HKE+EVY+DDMIVKS+
Sbjct: 183 EDMEKTTFITLWGTFCYKAMSFGLKNVGATYQRAMVALF*DMMHKEIEVYMDDMIVKSRT 362
Query: 709 EEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMP 768
EE+H+ L K+F RLRKY+LRLNP KC F V+S KLL FI S +GIEVD +KV+ I EM
Sbjct: 363 EEEHLVNLRKLFRRLRKYRLRLNPAKCMFEVKSRKLLDFIDS*RGIEVDSNKVKVILEMA 542
Query: 769 APQTEKQVRGFLRRLNYISRFISHMTATCGPIFKLLRKNQPIVWNDECQGAFDSIKNYLL 828
P TEKQV+GFL RLNYI RFIS + ATC P+F LL KNQ + W+ +C AF+ IK L+
Sbjct: 543 KPHTEKQVQGFLGRLNYIVRFIS*LIATCEPLFILLCKNQFVKWDHDC*VAFERIKQCLI 722
Query: 829 EPPILV 834
P +LV
Sbjct: 723 NPHVLV 740
>NP334778 reverse transcriptase [Glycine max]
Length = 431
Score = 221 bits (562), Expect = 3e-57
Identities = 101/143 (70%), Positives = 118/143 (81%)
Frame = +3
Query: 597 RMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSF 656
RMCVD+RDLN+ASPKDNFPLPHID+L+ N A +FSFMDGFSGYNQIKM+PED EKT+F
Sbjct: 3 RMCVDYRDLNRASPKDNFPLPHIDILMANMASFALFSFMDGFSGYNQIKMAPEDMEKTTF 182
Query: 657 ITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVEYL 716
IT WGTFCYKVM FGL N GATY R M LF DM+HKE+E YVD+MI KS+ EE+H+ L
Sbjct: 183 ITLWGTFCYKVMSFGLKNFGATYHRAMVALFQDMMHKEIEAYVDEMIAKSRMEEEHLVNL 362
Query: 717 TKMFERLRKYKLRLNPNKCTFGV 739
+F +LRKY+LRLNP KC FG+
Sbjct: 363 QNLFGQLRKYRLRLNPRKCVFGL 431
>NP595172 polyprotein [Glycine max]
Length = 4659
Score = 215 bits (547), Expect = 1e-55
Identities = 130/375 (34%), Positives = 197/375 (51%)
Frame = +1
Query: 534 EHRIPTKPECPPVRQKLRRTHPDMALKIKSEV*KQIDAGFLMTIEYPEWVANIVPVPKKD 593
+H IP K PV+ + R +I+ + + + G + P + I+ V KKD
Sbjct: 1756 DHAIPLKQGSGPVKVRPYRYPHTQKDQIEKMIQEMLVQGIIQPSNSP-FSLPILLVKKKD 1932
Query: 594 GKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREK 653
G R C D+R LN + KD+FP+P +D L+D ++ FS +D SGY+QI + PEDREK
Sbjct: 1933 GSWRFCTDYRALNAITVKDSFPMPTVDELLDELHGAQYFSKLDLRSGYHQILVQPEDREK 2112
Query: 654 TSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHV 713
T+F T G + + VMPFGL NA AT+Q M +F + K V V+ DD+++ S + H+
Sbjct: 2113 TAFRTHHGHYEWLVMPFGLTNAPATFQCLMNKIFQFALRKFVLVFFDDILIYSASWKDHL 2292
Query: 714 EYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQTE 773
++L + + L++++L +KC+FG LG VS G+ ++ KV+A+ + P P
Sbjct: 2293 KHLESVLQTLKQHQLFARLSKCSFGDTEVDYLGHKVSGLGVSMENTKVQAVLDWPTPNNV 2472
Query: 774 KQVRGFLRRLNYISRFISHMTATCGPIFKLLRKNQPIVWNDECQGAFDSIKNYLLEPPIL 833
KQ+RGFL Y RFI GP+ LL+K+ +WN+E + AF +K + E P+L
Sbjct: 2473 KQLRGFLGLTGYYRRFIKSYANIAGPLTDLLQKDS-FLWNNEAEAAFVKLKKAMTEAPVL 2649
Query: 834 VPPMEGKPLIMYLSVFDESVGCVLGQQDETGKKEHAIYYLSKKFTDCETRYMMLEKTCCA 893
P +P I+ VG VLGQ H I Y SKK + + A
Sbjct: 2650 SLPDFSQPFILETDASGIGVGAVLGQNG------HPIAYFSKKLAPRMQKQSAYTRELLA 2811
Query: 894 PAWAAKRLRHYLVNH 908
A + RHYL+ +
Sbjct: 2812 ITEALSKFRHYLLGN 2856
>TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (6%)
Length = 402
Score = 206 bits (523), Expect = 9e-53
Identities = 96/133 (72%), Positives = 114/133 (85%)
Frame = +2
Query: 632 FSFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMI 691
FSFMDGFSGYNQI M+ ED EKT+F+T WGTF Y+VM FGL N GATYQR M LFHDM+
Sbjct: 2 FSFMDGFSGYNQI*MAREDVEKTTFVTLWGTFSYRVMAFGLKNTGATYQRAMVALFHDMM 181
Query: 692 HKEVEVYVDDMIVKSKDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQ 751
HKE+EVYVDDMI KS+ E +H+ L K+F RL+KY+L+LNP KCTFGV+SGKLLGFIVSQ
Sbjct: 182 HKEIEVYVDDMIAKSRTETEHLVNLCKLFGRLQKYQLKLNPTKCTFGVKSGKLLGFIVSQ 361
Query: 752 KGIEVDPDKVRAI 764
KGIE+DP+KV+A+
Sbjct: 362 KGIEIDPEKVKAL 400
>NP395547 reverse transcriptase [Glycine max]
Length = 762
Score = 183 bits (464), Expect = 6e-46
Identities = 95/252 (37%), Positives = 142/252 (55%), Gaps = 18/252 (7%)
Frame = +1
Query: 561 IKSEV*KQIDAGFLMTIEYPEWVANIVPVPKKDGKV------------------RMCVDF 602
++ EV K ++AG + I WV+ + VPKK G RMC+D+
Sbjct: 1 VRKEVFKLLEAGLIYPISDSSWVSPVQVVPKKGGMTVVKNDRNELIPTRRVTRWRMCIDY 180
Query: 603 RDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPWGT 662
R LN+A+ KD++PLP +D ++ A+ + F+DG+SGYNQI + P+D+EKT+F P+
Sbjct: 181 RKLNEATRKDHYPLPFMDQMLKRLARQSFYRFLDGYSGYNQIAVDPQDQEKTAFTCPFSV 360
Query: 663 FCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVEYLTKMFER 722
F Y+ MPFGL NA T+QR M +F DM+ K +EV++DD + L K+ +R
Sbjct: 361 FAYRRMPFGLCNASTTFQRCMMAIFDDMVEKCIEVFMDDFSFFGASFGNCLANLEKVLQR 540
Query: 723 LRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQTEKQVRGFLRR 782
K L LN KC F V+ G +LG +S++GIEV +K+ I ++P P K + FL
Sbjct: 541 CEKSNLVLNWEKCHFMVQEGIVLGHKISKRGIEVVKEKLDVIDKLPPPVNVKGIHSFLGH 720
Query: 783 LNYISRFISHMT 794
+ + RFI T
Sbjct: 721 VGFYRRFIKDFT 756
>NP395548 reverse transcriptase [Glycine max]
Length = 762
Score = 174 bits (442), Expect = 2e-43
Identities = 90/252 (35%), Positives = 141/252 (55%), Gaps = 18/252 (7%)
Frame = +1
Query: 561 IKSEV*KQIDAGFLMTIEYPEWVANIVPVPKKDGKV------------------RMCVDF 602
++ EV K ++ G + I WV+ ++ V KK+G ++C+D+
Sbjct: 1 VRKEVLKLLEVGLIYPISDSAWVSPVLVVSKKEGMTVIRNEKNDLIPTRTVTSWKLCIDY 180
Query: 603 RDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPWGT 662
R LN+A+ KD+FPLP +D +++ A + F+D + GYNQI + P+D+EK +F P+G
Sbjct: 181 RKLNEATRKDHFPLPFMDQMLERLAGHAYYCFLDAYFGYNQIVVDPKDQEKMAFTCPFGV 360
Query: 663 FCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVEYLTKMFER 722
F Y+ +PFGL NA T+Q M +F D++ K +EV++DD V E ++ L + +R
Sbjct: 361 FAYRRIPFGLCNAPTTFQMCMLAIFADIVEKSIEVFMDDFSVFVPSLESCLKKLEMVLQR 540
Query: 723 LRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPQTEKQVRGFLRR 782
+ L LN KC F VR G +LG +S +GIEVD K+ I ++P P K +R FL +
Sbjct: 541 CVETNLVLNWEKCHFMVREGIVLGHKISTRGIEVDQTKIDVIEKLPPPSNVKGIRSFLGQ 720
Query: 783 LNYISRFISHMT 794
+ RFI T
Sbjct: 721 ARFYRRFIKDFT 756
>TC219643 weakly similar to UP|Q6WAY7 (Q6WAY7) Gag/pol polyprotein
(Fragment), partial (8%)
Length = 1320
Score = 166 bits (420), Expect = 8e-41
Identities = 80/169 (47%), Positives = 111/169 (65%), Gaps = 2/169 (1%)
Frame = +3
Query: 424 RLSHSKPVGHSNPTFPLNFEFPVYEAEDEEGDDI--PYEITRLLEQERKAIQPHQEEIEL 481
R P G + ++FE + + EDE +D+ P E+ R++ E + + PHQEE EL
Sbjct: 813 RSDDESPEGTNTWDPSIDFEQKMNQTEDEGNEDVGLPPELERMVAHEDQEMGPHQEETEL 992
Query: 482 INLGTEENKREIKVGAALEEGVKRKIFQLL*EYPDIFAWSYEDMPGLDPKIVEHRIPTKP 541
++LG+ KRE+K+G + ++ ++ LL +Y DIFAWSY+DMPGL IV+HR+P P
Sbjct: 993 VDLGSGSGKREVKIGTGITAPIREELIILLRDYQDIFAWSYQDMPGLSSDIVQHRLPLNP 1172
Query: 542 ECPPVRQKLRRTHPDMALKIKSEV*KQIDAGFLMTIEYPEWVANIVPVP 590
EC PV+QKLRR P+ +LKIK EV K DAGFL YP+WVANIVP+P
Sbjct: 1173ECSPVKQKLRRMKPETSLKIKEEVKK*FDAGFLAVARYPKWVANIVPIP 1319
>BQ628592
Length = 423
Score = 100 bits (248), Expect(2) = 3e-38
Identities = 44/76 (57%), Positives = 58/76 (75%)
Frame = -1
Query: 804 LRKNQPIVWNDECQGAFDSIKNYLLEPPILVPPMEGKPLIMYLSVFDESVGCVLGQQDET 863
L KNQ I+WN Q AF+ IK L P +L+PP+ G+P ++Y+++ DES+GCVL Q D++
Sbjct: 423 LPKNQAILWNSNYQEAFEKIKQSLANPSVLMPPVTGRPFLLYMTMLDESMGCVLVQHDDS 244
Query: 864 GKKEHAIYYLSKKFTD 879
GKKE AIYYLSKKFTD
Sbjct: 243 GKKEQAIYYLSKKFTD 196
Score = 79.0 bits (193), Expect(2) = 3e-38
Identities = 34/50 (68%), Positives = 42/50 (84%)
Frame = -3
Query: 884 YMMLEKTCCAPAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKPAVLLGRL 933
Y MLE+TCC WA+ RLR Y+++HTTWLIS+MDP+KYIFEKPA L GR+
Sbjct: 184 YSMLERTCCTLVWASHRLRQYMLSHTTWLISKMDPVKYIFEKPA-LTGRI 38
>BG839293
Length = 781
Score = 112 bits (281), Expect = 1e-24
Identities = 59/147 (40%), Positives = 88/147 (59%), Gaps = 2/147 (1%)
Frame = +1
Query: 404 Y*FQKICIKTSF*NHPTILRRLSHSKPVGHSNPTFPLNFEFPVYEAEDEEGDDI--PYEI 461
Y F IC+ TS + R++ P G + +NFE + EDE +D+ P E+
Sbjct: 301 YPFSFICMFTSL----FLKRQI**RVPEGTNTWDPLINFEQETSQTEDEGNEDVGLPPEL 468
Query: 462 TRLLEQERKAIQPHQEEIELINLGTEENKREIKVGAALEEGVKRKIFQLL*EYPDIFAWS 521
R++ E + + PHQEE EL++LG KRE+K+G + ++ ++ LL +Y DIFAWS
Sbjct: 469 ERMVAHEDQEMGPHQEETELVDLGIGSGKREVKIGTGITAPIREELIILLKDYQDIFAWS 648
Query: 522 YEDMPGLDPKIVEHRIPTKPECPPVRQ 548
Y+DMPGL IV+H++P PEC PV+Q
Sbjct: 649 YQDMPGLSSDIVQHQLPLNPECSPVKQ 729
>TC223727 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (9%)
Length = 843
Score = 107 bits (268), Expect = 3e-23
Identities = 77/232 (33%), Positives = 121/232 (51%)
Frame = +3
Query: 1170 R*TMVL*HQVVLAKP*ISTGCLQQGQEDPEKIGQ*IPVRWRYSVQEKLRHGIVKIC**TR 1229
R* +V HQ + K I +Q ED EK+G +++V EK RH +C
Sbjct: 129 R*ALVFRHQAICHKQRIPARDCRQ**EDIEKVGGRFLHERKHTV*EKPRHETSAVCGCQG 308
Query: 1230 SGAVNA*CT*RYLREPRYGAHYAKKVVTSRLLLDDHGA*LLPTLQKVP*MLNLC**DSCA 1289
+ + * + R A Y ++ SRLLL HG *LL +++P M ++ * C
Sbjct: 309 GKSHDRGSP*GLVWNARQRACYGQEDPKSRLLLAYHGK*LLCPCEEMPQMSSVRR*CQCP 488
Query: 1290 SACPQCYLLPMAVLHVGHRHDWKN*TASIKWSSFHFSGN*LLHQMG*SGILYQCNQASGS 1349
+A +C++LP+A LHVG+R +* ++WSS H + L HQ+G LY+ ++ G
Sbjct: 489 TASSECHVLPLAFLHVGNRCHRGH*AQGLEWSSLHPRSDRLFHQVGRGSFLYRRHEGCGG 668
Query: 1350 QVHQEQHHLSIWCSQQDHYRKWYQPEQQCGASSL*RIQDRTS*FFSL*ASDE 1401
QVH E+ HL IW +++D++ + +QP+ Q +L + + S L A DE
Sbjct: 669 QVH*ERDHLPIWFAKEDYHGQRHQPK*QDDGGNLRGV*NPASQSHXLPAKDE 824
>TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%)
Length = 589
Score = 53.1 bits (126), Expect(2) = 9e-12
Identities = 43/171 (25%), Positives = 69/171 (40%)
Frame = +1
Query: 765 REMPAPQTEKQVRGFLRRLNYISRFISHMTATCGPIFKLLRKNQPIVWNDECQGAFDSIK 824
R P ++ +R F ++ RF+ + + P+ +L++KN W ++ + AF +K
Sbjct: 88 RMAPTLKSVGDIRSFHGLASFYRRFVPNFSTVASPLNELVKKNMAFTWGEKQEQAFALLK 267
Query: 825 NYLLEPPILVPPMEGKPLIMYLSVFDESVGCVLGQQDETGKKEHAIYYLSKKFTDCETRY 884
L + P+L P K + V VL Q H I Y S+K Y
Sbjct: 268 EKLTKAPVLALPDFSKTFELECDASGVGVRAVLLQGG------HPIAYFSEKLHSATLNY 429
Query: 885 MMLEKTCCAPAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKPAVLLGRLHA 935
+K A A + H+LV + S +KYI K L + HA
Sbjct: 430 PTYDKELYALIRAPQTWEHFLVCKEFVIHSDHQSLKYIRGKSK--LNKRHA 576
Score = 36.6 bits (83), Expect(2) = 9e-12
Identities = 12/25 (48%), Positives = 21/25 (84%)
Frame = +2
Query: 746 GFIVSQKGIEVDPDKVRAIREMPAP 770
GF+V + G+++DP+K++AI+E P P
Sbjct: 29 GFVVGRNGVQMDPEKIKAIQEWPPP 103
>TC224482 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (6%)
Length = 669
Score = 69.7 bits (169), Expect = 1e-11
Identities = 57/181 (31%), Positives = 86/181 (47%), Gaps = 2/181 (1%)
Frame = +3
Query: 1408 Q*EHQEDCPENGNYLQRLA*DVALCSAWLPYYSA*FNRGNPFLSCIWYGSSTSF-GSGDL 1466
Q*E+Q+D P++ +Q LA D + LP +SA N GN L IW G + G +
Sbjct: 9 Q*EYQKDYPKDDRVIQGLARDAPIRVTRLPDFSANVNWGNAVLIGIWDGGCVTV*GRSPV 188
Query: 1467 ISPCDHGSKVIRG*MV-PKPV*SVELG*GKTYGCRGSWTVISSKNEDCL*QESPSSRIQG 1525
I D G I+G V +*S + *G SW ++ +KNE+C+ QE +++
Sbjct: 189 IK--DLGRIRIKGIRVGSNAL*SAQPH*G*ALNGHESWALVPAKNEECIRQEGMLAQVP* 362
Query: 1526 R*TCIEKEDKPTTRPKGQVDA*L*GSLCCQEGLLWWCFNSYTHGWCRTSKSCEC*YSQEI 1585
C ++ P ++ L + CC+EG +Y HGW R + + E * Q I
Sbjct: 363 GRPCAKENVPCCQGPSREMGPELRRAFCCEEGFFRRSPGAYQHGW*RVTFTHEF*CRQAI 542
Query: 1586 L 1586
L
Sbjct: 543 L 545
>AW570005
Length = 413
Score = 65.5 bits (158), Expect = 2e-10
Identities = 43/141 (30%), Positives = 61/141 (42%)
Frame = -2
Query: 768 PAPQTEKQVRGFLRRLNYISRFISHMTATCGPIFKLLRKNQPIVWNDECQGAFDSIKNYL 827
P P+T + +RGFLR + RFI A P+ LL K+ VW+ E AF ++KN +
Sbjct: 412 PPPRTARSLRGFLRLTGFYRRFIKGYAAMAAPLSHLLTKDS-FVWSPEADVAFQALKNVV 236
Query: 828 LEPPILVPPMEGKPLIMYLSVFDESVGCVLGQQDETGKKEHAIYYLSKKFTDCETRYMML 887
+L P KP + +G VL Q+ H I + SK+F R
Sbjct: 235 TNTLVLALPDFTKPFTVETDASGSDMGAVLSQEG------HPIAFFSKEFCPKLVRSSTY 74
Query: 888 EKTCCAPAWAAKRLRHYLVNH 908
A K+ R YL+ H
Sbjct: 73 VHELAAITNVVKKWRQYLLGH 11
>BI317507
Length = 359
Score = 65.1 bits (157), Expect = 2e-10
Identities = 34/114 (29%), Positives = 59/114 (50%)
Frame = -1
Query: 701 DMIVKSKDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDK 760
++++ S D + H+ +LT + + L+K +L N KC F + + LG ++S+ + +D +K
Sbjct: 353 NILIYSPDWKSHIMHLTAVLDVLKKERLVANRKKCYFSQTTIEYLGHVISKDCVAMDSNK 174
Query: 761 VRAIREMPAPQTEKQVRGFLRRLNYISRFISHMTATCGPIFKLLRKNQPIVWND 814
V+++ E P P+ K+V FLR Y +FI L KN W D
Sbjct: 173 VKSVIEWPVPKNVKRVCSFLRLTGYYRKFIKDYGKLAPRPLTDLTKNDGFKWGD 12
>BE802896
Length = 416
Score = 59.3 bits (142), Expect = 1e-08
Identities = 37/128 (28%), Positives = 58/128 (44%)
Frame = -2
Query: 779 FLRRLNYISRFISHMTATCGPIFKLLRKNQPIVWNDECQGAFDSIKNYLLEPPILVPPME 838
FL + RFI P+ LL+K +ND+C+ AFD +K L+ PI+ P
Sbjct: 412 FLGHAGFYRRFIRDFRKVALPLSNLLQKEVEFDFNDKCK*AFDCLKRALITTPIIQAPDW 233
Query: 839 GKPLIMYLSVFDESVGCVLGQQDETGKKEHAIYYLSKKFTDCETRYMMLEKTCCAPAWAA 898
P + + ++G VL Q+ + K IYY S+ + Y EK A +A
Sbjct: 232 TAPFELMCDASNYALGVVLAQKID--KLPRVIYYSSRTLDAAQANYTTTEKELLAIVFAL 59
Query: 899 KRLRHYLV 906
++ YL+
Sbjct: 58 EKFHSYLL 35
>BG725601 similar to PIR|H86337|H86 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (1%)
Length = 285
Score = 58.9 bits (141), Expect = 2e-08
Identities = 32/90 (35%), Positives = 49/90 (53%)
Frame = -3
Query: 535 HRIPTKPECPPVRQKLRRTHPDMALKIKSEV*KQIDAGFLMTIEYPEWVANIVPVPKKDG 594
H++ + V Q+ R+ + + EV K A F+ I Y + ++V V K +G
Sbjct: 277 HKLAICNDVKLVTQRKRKIREERCQTV*QEVVKLAIASFIRDINYST*LFSVVMVKKPNG 98
Query: 595 KVRMCVDFRDLNKASPKDNFPLPHIDVLVD 624
K R+C D+ DLN A PKD +PLP+ID + D
Sbjct: 97 KWRICTDYIDLN*ACPKDAYPLPNIDHMTD 8
>AW184779
Length = 432
Score = 47.4 bits (111), Expect = 5e-05
Identities = 25/52 (48%), Positives = 32/52 (61%), Gaps = 5/52 (9%)
Frame = +2
Query: 1298 LPMAVL-----HVGHRHDWKN*TASIKWSSFHFSGN*LLHQMG*SGILYQCN 1344
+P+ VL +VGHR D +* KW+S HFS N*LLHQMG S + C+
Sbjct: 257 IPLNVLAAPWPYVGHRRDRSH*AQGFKWTSLHFSRN*LLHQMGRSSFVC*CD 412
>BI498328
Length = 335
Score = 39.3 bits (90), Expect = 0.014
Identities = 21/44 (47%), Positives = 28/44 (62%)
Frame = +3
Query: 1019 RNRGNRCIPAGAPYSFYRPDSVRMYKQYGRIRSMYLWDRGSN*H 1062
R+RG+ CIP + YSF+ +Y+QYGR+RSM RGS H
Sbjct: 192 RSRGSPCIPG*SVYSFHG*TRF*LYQQYGRVRSMR--PRGSGGH 317
>BI785135
Length = 420
Score = 38.5 bits (88), Expect = 0.024
Identities = 17/71 (23%), Positives = 37/71 (51%)
Frame = +1
Query: 753 GIEVDPDKVRAIREMPAPQTEKQVRGFLRRLNYISRFISHMTATCGPIFKLLRKNQPIVW 812
G+ ++P +++ I E P P + + + GF N+ RF+ + P+ +L+R + P W
Sbjct: 208 GVPMNPKRIKVIPEWPTPPSIR*IWGFHNLTNFYKRFVP*FSILVAPLIELVRNHVP-SW 384
Query: 813 NDECQGAFDSI 823
+ + F ++
Sbjct: 385 EEAQEMGFQTL 417
>BE804087
Length = 160
Score = 37.0 bits (84), Expect = 0.069
Identities = 22/51 (43%), Positives = 29/51 (56%)
Frame = +1
Query: 1407 SQ*EHQEDCPENGNYLQRLA*DVALCSAWLPYYSA*FNRGNPFLSCIWYGS 1457
SQ*E+ E E+ + +QRLA*DV C A++ F GN L +W GS
Sbjct: 7 SQ*EY*EKYLEDDSVVQRLA*DVFFCPAYVSNLGTNFYWGNTLLLGLWEGS 159
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.346 0.153 0.549
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 80,395,057
Number of Sequences: 63676
Number of extensions: 1340901
Number of successful extensions: 12423
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 8961
Number of HSP's successfully gapped in prelim test: 334
Number of HSP's that attempted gapping in prelim test: 2948
Number of HSP's gapped (non-prelim): 10014
length of query: 1586
length of database: 12,639,632
effective HSP length: 110
effective length of query: 1476
effective length of database: 5,635,272
effective search space: 8317661472
effective search space used: 8317661472
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.7 bits)
S2: 65 (29.6 bits)
Medicago: description of AC147714.3


