

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147712.15 + phase: 0 /pseudo/partial
(740 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
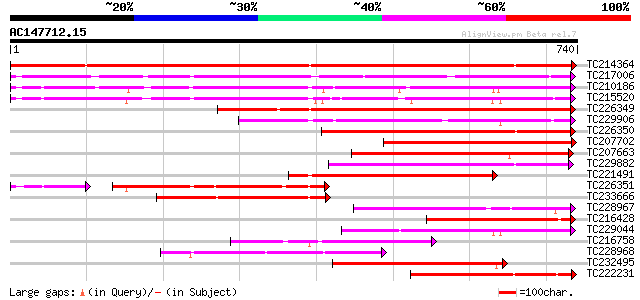
Score E
Sequences producing significant alignments: (bits) Value
TC214364 UP|Q6WNU4 (Q6WNU4) Subtilisin-like protease, complete 829 0.0
TC217006 UP|Q9ZTT3 (Q9ZTT3) Subtilisin-like protease C1, complete 510 e-144
TC210186 UP|Q93WQ0 (Q93WQ0) Subtilisin-type protease, complete 485 e-137
TC215520 GB|AAG38994.1|11611651|AF160513 subtilisin-type proteas... 434 e-122
TC226349 similar to PIR|JC7519|JC7519 subtilisin-like serine pro... 353 2e-97
TC229906 weakly similar to UP|Q9ZTT3 (Q9ZTT3) Subtilisin-like pr... 269 3e-72
TC226350 similar to PIR|JC7519|JC7519 subtilisin-like serine pro... 268 6e-72
TC207702 weakly similar to UP|Q8S896 (Q8S896) Subtilisin-like se... 265 4e-71
TC207663 similar to UP|P93204 (P93204) SBT1 protein (Subtilisin-... 262 4e-70
TC229882 weakly similar to UP|Q75I27 (Q75I27) Putaive subtilisin... 235 5e-62
TC221491 similar to UP|P93204 (P93204) SBT1 protein (Subtilisin-... 234 8e-62
TC226351 similar to PIR|JC7519|JC7519 subtilisin-like serine pro... 212 1e-61
TC233666 weakly similar to UP|Q9FJF3 (Q9FJF3) Serine protease-li... 223 2e-58
TC228967 similar to GB|BAB09208.1|9758669|AB012245 subtilisin-li... 220 2e-57
TC216428 similar to UP|Q9AX30 (Q9AX30) Subtilisin-like protease,... 220 2e-57
TC229044 similar to UP|Q93WQ0 (Q93WQ0) Subtilisin-type protease,... 219 3e-57
TC216758 similar to PIR|T05768|T05768 subtilisin-like proteinase... 216 2e-56
TC228968 similar to GB|BAB09208.1|9758669|AB012245 subtilisin-li... 205 5e-53
TC232495 similar to UP|Q8W554 (Q8W554) AT3g14240/MLN21_2, partia... 203 3e-52
TC222231 weakly similar to UP|Q9FVC7 (Q9FVC7) Subtilase , partia... 184 9e-47
>TC214364 UP|Q6WNU4 (Q6WNU4) Subtilisin-like protease, complete
Length = 2667
Score = 829 bits (2141), Expect = 0.0
Identities = 415/743 (55%), Positives = 528/743 (70%), Gaps = 4/743 (0%)
Frame = +1
Query: 1 SYIVYIGSHSHGPNPSASDLQSATDSHYNLLGSHLGSHEKAKEAIFYSYNKHINGFAAVL 60
SY+VY+G+HSHGP S+ D T SH++ LGS LGS AK++IFYSY +HINGFAA L
Sbjct: 124 SYVVYLGAHSHGPELSSVDFNQVTQSHHDFLGSFLGSSNTAKDSIFYSYTRHINGFAATL 303
Query: 61 EVEEAAKIAKHPNVVSVFENKGHELQTTRSWEFLGLENNYGVVPKDSIWEKGRYGEGTII 120
+ E A +IAKHP V+SVFEN+G +L TTRSW+F+ LE+N GV+ SIW+K R+GEG II
Sbjct: 304 DEEVAVEIAKHPKVLSVFENRGRKLHTTRSWDFMELEHN-GVIQSSSIWKKARFGEGVII 480
Query: 121 ANIDSGVSPESKSFSDDGMGPVPSRWRGICQ--LDN-FHCNRKLIGARFYSQGYESKFGR 177
N+D+GV PESKSFS+ G+GP+PS+WRGIC +D+ FHCNRKLIGAR++++GY S G
Sbjct: 481 GNLDTGVWPESKSFSEQGLGPIPSKWRGICHNGIDHTFHCNRKLIGARYFNKGYASVAGP 660
Query: 178 LNQSLYNARDVLGHGTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVCWLGTI 237
LN S + RD GHGT TLS AGGN V+ +VFG +GTAKGGSP + VAAYKVCW
Sbjct: 661 LNSSFDSPRDNEGHGTHTLSTAGGNMVARVSVFGQGHGTAKGGSPMARVAAYKVCWPPVA 840
Query: 238 *IECTDADIMQAFEDAISDGVDIISCSLGQTSPKEFFEDGISIGAFHAIENGVIVVAGGG 297
EC DADI+ AF+ AI DGVD++S SLG +S FF+D ++IG+FHA + GV+VV G
Sbjct: 841 GDECFDADILAAFDLAIHDGVDVLSVSLGGSS-STFFKDSVAIGSFHAAKRGVVVVCSAG 1017
Query: 298 NSGPKFGTVTNVAPWLFSVAASTIDRNFVSYLQLGDKHIIMGTSLSTGLPNEKFYSLVSS 357
NSGP T N+APW +VAAST+DR F +Y+ LG+ G SLS KFY ++ +
Sbjct: 1018NSGPAEATAENLAPWHVTVAASTMDRQFPTYVVLGNDITFKGESLSATKLAHKFYPIIKA 1197
Query: 358 VDAKVGNATIEDAKICKVGSLDPNKVKGKILFCLLRELDGLVYAEEEAISGGSIGLVLGN 417
DAK+ +A EDA +C+ G+LDPNK KGKI+ CL R ++ V E+A G++G+VL N
Sbjct: 1198TDAKLASARAEDAVLCQNGTLDPNKAKGKIVVCL-RGINARVDKGEQAFLAGAVGMVLAN 1374
Query: 418 DKQRGNDIMAYAHLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAKTEVGVKPAPVIASL 477
DK GN+I+A H+LP SHIN+TDG V YI +TK P+AY+T KT++ KPAP +A+
Sbjct: 1375DKTTGNEIIADPHVLPASHINFTDGSAVFKYINSTKFPVAYITHPKTQLDTKPAPFMAAF 1554
Query: 478 SSRGPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISCPHVS 537
SS+GPN I P ILKPDITAPGV ++ AY A PT D + IP+N SGTS+SCPHVS
Sbjct: 1555SSKGPNTIVPEILKPDITAPGVSVIAAYTEAQGPTNQVFDKRRIPFNSVSGTSMSCPHVS 1734
Query: 538 AIVALLKTIYPNWSPAAFKSAIMTTTTIQGNNHRPIKDQSKEDATPFGYGAGHIQPELAM 597
IV LL+ +YP WS AA KSAIMTT T N P+ + + ATPF YGAGH+QP AM
Sbjct: 1735GIVGLLRALYPTWSTAAIKSAIMTTATTLDNEVEPLLNATDGKATPFSYGAGHVQPNRAM 1914
Query: 598 DPGLVYDLNIVDYLNFLCAHGYNQTQMKMFSRKPYICPKSYNMLDFNYPSITVPNLGKHF 657
DPGLVYD+ I DYLNFLCA GYN+TQ+ +F+ PY C K +++L+ NYPSITVP L
Sbjct: 1915DPGLVYDITIDDYLNFLCALGYNETQISVFTEGPYKCRKKFSLLNLNYPSITVPKLSGSV 2094
Query: 658 VQEVTRTVTNVGSPGTYRVQVNEPHGIFVLIKPRSLTFNEVGEKKTFKIIFKVTK-PTSS 716
VTRT+ NVGSPGTY V P+GI V +KP L F VGE+K+FK+ FK + ++
Sbjct: 2095T--VTRTLKNVGSPGTYIAHVQNPYGITVSVKPSILKFKNVGEEKSFKLTFKAMQGKATN 2268
Query: 717 GYVFGHLLWSDGRHKVMSPLVVK 739
Y FG L+WSDG+H V SP+VVK
Sbjct: 2269NYAFGKLIWSDGKHYVTSPIVVK 2337
>TC217006 UP|Q9ZTT3 (Q9ZTT3) Subtilisin-like protease C1, complete
Length = 2422
Score = 510 bits (1313), Expect = e-144
Identities = 306/749 (40%), Positives = 428/749 (56%), Gaps = 11/749 (1%)
Frame = +3
Query: 1 SYIVYIGSHSHGPNPSASDLQSATDSHYNLLGSHLGSHEKAKEAIFYSYNKHINGFAAVL 60
SYIVY G+ S +D SA + ++L S+ + K + + + + +GF A+L
Sbjct: 117 SYIVYTGN-------SMNDEASALTLYSSMLQEVADSNAEPK-LVQHHFKRSFSGFVAML 272
Query: 61 EVEEAAKIAKHPNVVSVFENKGHELQTTRSWEFLGLENNYGVVPKDSIWEKGRYGEGTII 120
EEA ++A+H VV+VF NK +L TTRSW+F+G P +S II
Sbjct: 273 TEEEADRMARHDRVVAVFPNKKKQLHTTRSWDFIGFPLQANRAPAES---------DVII 425
Query: 121 ANIDSGVSPESKSFSDDGMGPVPSRWRGICQLD-NFHCNRKLIGARFYS-QGYESKFGRL 178
A DSG+ PES+SF+D G GP PS+W+G CQ NF CN K+IGA+ Y G+ SK
Sbjct: 426 AVFDSGIWPESESFNDKGFGPPPSKWKGTCQTSKNFTCNNKIIGAKIYKVDGFFSK---- 593
Query: 179 NQSLYNARDVLGHGTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVCWLGTI* 238
+ RD+ GHGT S A GN VS A++ GL GT++GG ++ +A YKVCW
Sbjct: 594 -DDPKSVRDIDGHGTHVASTAAGNPVSTASMLGLGQGTSRGGVTKARIAVYKVCWFDG-- 764
Query: 239 IECTDADIMQAFEDAISDGVDIISCSLGQTSPKEFFEDGISIGAFHAIENGVIVVAGGGN 298
CTDADI+ AF+DAI+DGVDII+ SLG S + +F DGI+IGAFHA+ NGV+ V GN
Sbjct: 765 --CTDADILAAFDDAIADGVDIITVSLGGFSDENYFRDGIAIGAFHAVRNGVLTVTSAGN 938
Query: 299 SGPKFGTVTNVAPWLFSVAASTIDRNFVSYLQLGDKHIIMGTSLSTGLPNEKFYSLVSSV 358
SGP+ +++N +PW SVAASTIDR FV+ ++LG+K GTS++T + Y ++
Sbjct: 939 SGPRPSSLSNFSPWSISVAASTIDRKFVTKVELGNKITYEGTSINTFDLKGELYPIIYGG 1118
Query: 359 DAKVGNATIE--DAKICKVGSLDPNKVKGKILFCLLRELDGLVYAEEEAISGGSIGLVLG 416
DA I+ ++ C GSLD VKGKI+ C R +A+ G V
Sbjct: 1119DAPNKGEGIDGSSSRYCSSGSLDKKLVKGKIVLCESR---------SKALGPFDAGAVGA 1271
Query: 417 NDKQRGNDIMAYAHLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAKTEVGVKPAPVIAS 476
+ +G + + LP S++ DG V+ YI +T+TP+A + K E APV+AS
Sbjct: 1272LIQGQGFRDLPPSLPLPGSYLALQDGASVYDYINSTRTPIATIFKT-DETKDTIAPVVAS 1448
Query: 477 LSSRGPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISCPHV 536
SSRGPN + P ILKPD+ APGV IL ++ A P+ + DN+ + +NI SGTS++CPHV
Sbjct: 1449FSSRGPNIVTPEILKPDLVAPGVSILASWSPASPPSDVEGDNRTLNFNIISGTSMACPHV 1628
Query: 537 SAIVALLKTIYPNWSPAAFKSAIMTTT-TIQGNNHRPIKDQSKEDATPFGYGAGHIQPEL 595
S A +K+ +P WSPAA +SA+MTT + H F YGAG I P
Sbjct: 1629SGAAAYVKSFHPTWSPAAIRSALMTTAKQLSPKTHL---------LAEFAYGAGQIDPSK 1781
Query: 596 AMDPGLVYDLNIVDYLNFLCAHGYNQTQMKMFSRKPYICPKSYN--MLDFNYPS--ITVP 651
A+ PGLVYD +DY+ FLC GY+ +++ + CP++ N D NY S + VP
Sbjct: 1782AVYPGLVYDAGEIDYVRFLCGQGYSTRTLQLITGDNSSCPETKNGSARDLNYASFALFVP 1961
Query: 652 NLGKHFVQ-EVTRTVTNVGSP-GTYRVQVNEPHGIFVLIKPRSLTFNEVGEKKTFKIIFK 709
+ V RTVTNVGSP TY+ V P G+ + + P L F + +K+TF +
Sbjct: 1962PYNSNSVSGSFNRTVTNVGSPKSTYKATVTSPKGLKIEVNPSVLPFTSLNQKQTF--VLT 2135
Query: 710 VTKPTSSGYVFGHLLWSDGRHKVMSPLVV 738
+T V G L+W DG+++V SP+VV
Sbjct: 2136ITGKLEGPIVSGSLVWDDGKYQVRSPIVV 2222
>TC210186 UP|Q93WQ0 (Q93WQ0) Subtilisin-type protease, complete
Length = 2401
Score = 485 bits (1248), Expect = e-137
Identities = 301/759 (39%), Positives = 429/759 (55%), Gaps = 22/759 (2%)
Frame = +1
Query: 2 YIVYIGSHSHGPNPSASDLQSATDSHYNLLGSHLGSHEKAKEAIFYSYNKHINGFAAVLE 61
YIVY+G+ + S S + H +L S L +E A + +Y +GFAA L
Sbjct: 181 YIVYMGA-------ADSTDASFRNDHAQVLNSVLRRNENA---LVRNYKHGFSGFAARLS 330
Query: 62 VEEAAKIAKHPNVVSVFENKGHELQTTRSWEFLGLENNYGVVPKDSIWEKGRYGEGTIIA 121
+EA IA+ P VVSVF +L TTRSW+FL + + K + K ++I
Sbjct: 331 KKEATSIAQKPGVVSVFPGPVLKLHTTRSWDFLKYQTQVKIDTKPNAVSKS----SSVIG 498
Query: 122 NIDSGVSPESKSFSDDGMGPVPSRWRGICQLD----NFHCNRKLIGARFYSQGYESKFGR 177
+D+G+ PE+ SFSD GMGPVPSRW+G C + +CNRKLIGAR+Y+ +S
Sbjct: 499 ILDTGIWPEAASFSDKGMGPVPSRWKGTCMKSQDFYSSNCNRKLIGARYYADPNDSGDN- 675
Query: 178 LNQSLYNARDVLGHGTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVCWLGTI 237
ARD GHGT A G V+ A+ +G+A G AKGGSP S +A Y+VC
Sbjct: 676 ------TARDSNGHGTHVAGTAAGVMVTNASYYGVATGCAKGGSPESRLAVYRVCSN--- 828
Query: 238 *IECTDADIMQAFEDAISDGVDIISCSLGQTSP--KEFFEDGISIGAFHAIENGVIVVAG 295
C + I+ AF+DAI+DGVD++S SLG ++ + D IS+GAFHA+E+G++VV
Sbjct: 829 -FGCRGSSILAAFDDAIADGVDLLSVSLGASTGFRPDLTSDPISLGAFHAMEHGILVVCS 1005
Query: 296 GGNSGPKFGTVTNVAPWLFSVAASTIDRNFVSYLQLGDKHIIMGTSLSTG-LPNEKFYSL 354
GN GP T+ N APW+ +VAASTIDRNF+S + LGD II G +++ L N Y L
Sbjct: 1006AGNDGPSSYTLVNDAPWILTVAASTIDRNFLSNIVLGDNKIIKGKAINLSPLSNSPKYPL 1185
Query: 355 VSSVDAKVGNATIEDAKICKVGSLDPNKVKGKILFCLLRELDGLVYAEEEAISG----GS 410
+ AK + ++ +A+ C SLD NKVKGKI+ C + Y+ + ++ G
Sbjct: 1186IYGESAKANSTSLVEARQCHPNSLDGNKVKGKIVVC---DDKNDKYSTRKKVATVKAVGG 1356
Query: 411 IGLVLGNDKQRGNDIMAYAHLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAKTEVGVKP 470
IGLV D+ I + P + I+ DG + YI +T P+A + + + KP
Sbjct: 1357IGLVHITDQNEA--IASNYGDFPATVISSKDGVTILQYINSTSNPVATILATTSVLDYKP 1530
Query: 471 APVIASLSSRGPNPIQPIILKPDITAPGVDILYAYIG---AISPTGLASDNQWIPYNIGS 527
AP++ + SSRGP+ + ILKPDI APGV+IL A+IG + P G Y I S
Sbjct: 1531APLVPNFSSRGPSSLSSNILKPDIAAPGVNILAAWIGNGTEVVPKGKKPSL----YKIIS 1698
Query: 528 GTSISCPHVSAIVALLKTIYPNWSPAAFKSAIMTTTTIQGNNHRPIKDQSKEDATPFGYG 587
GTS++CPHVS + + +KT P WS ++ KSAIMT+ N PI +S ATP+ YG
Sbjct: 1699GTSMACPHVSGLASSVKTRNPTWSASSIKSAIMTSAIQSNNLKAPITTESGSVATPYDYG 1878
Query: 588 AGHIQPELAMDPGLVYDLNIVDYLNFLCAHGYNQTQMKMFSR---KPYICPK---SYNML 641
AG + + PGLVY+ + VDYLNFLC G+N T +K+ S+ + + CPK S ++
Sbjct: 1879AGEMTTSEPLQPGLVYETSSVDYLNFLCYIGFNVTTVKVISKTVPRNFNCPKDLSSDHIS 2058
Query: 642 DFNYPSITVPNLGKHFVQEVTRTVTNVGSPG--TYRVQVNEPHGIFVLIKPRSLTFNEVG 699
+ NYPSI + GK V ++RTVTNVG Y V+ P G+ V + P L F +
Sbjct: 2059NINYPSIAINFSGKRAV-NLSRTVTNVGEDDETVYSPIVDAPSGVHVTLTPNKLRFTKSS 2235
Query: 700 EKKTFKIIFKVTKPTSSGYVFGHLLWSDGRHKVMSPLVV 738
+K ++++IF T + +FG + WS+G++ V SP V+
Sbjct: 2236KKLSYRVIFSSTLTSLKEDLFGSITWSNGKYMVRSPFVL 2352
>TC215520 GB|AAG38994.1|11611651|AF160513 subtilisin-type protease precursor
{Glycine max;} , complete
Length = 2392
Score = 434 bits (1115), Expect = e-122
Identities = 298/768 (38%), Positives = 409/768 (52%), Gaps = 31/768 (4%)
Frame = +2
Query: 2 YIVYIGSHSHGPNPSASDLQSATDSHYNLLGSHLGSHEKAKEAIFYSYNKHINGFAAVLE 61
YIVY+G+ + S S + H +L S L +E A + +Y +GFAA L
Sbjct: 149 YIVYMGA-------ADSTKASLKNEHAQILNSVLRRNENA---LVRNYKHGFSGFAARLS 298
Query: 62 VEEAAKIAKHPNVVSVFENKGHELQTTRSWEFLGLENNYGVVPKDSIWEKGRYGEGTIIA 121
EEA IA+ P VVSVF + +L TTRSW+FL + + K + + +I
Sbjct: 299 KEEANSIAQKPGVVSVFPDPILKLHTTRSWDFLKSQTRVNIDTKPNTLSGSSFSSSDVIL 478
Query: 122 NI-DSGVSPESKSFSDDGMGPVPSRWRGICQ----LDNFHCNRKLIGARFYSQGYESKFG 176
+ D+G+ PE+ SFSD G GPVPSRW+G C ++ CNRK+IGARFY E
Sbjct: 479 GVLDTGIWPEAASFSDKGFGPVPSRWKGTCMTSKDFNSSCCNRKIIGARFYPNPEEK--- 649
Query: 177 RLNQSLYNARDVLGHGTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVCWLGT 236
ARD GHGT S A G VSGA+ +GLA GTA+GGSP S +A YKVC
Sbjct: 650 -------TARDFNGHGTHVSSTAVGVPVSGASFYGLAAGTARGGSPESRLAVYKVCGAFG 808
Query: 237 I*IECTDADIMQAFEDAISDGVDIISCSLGQT--SPKEFFEDGISIGAFHAIENGVIVVA 294
C + I+ F+DAI DGVDI+S SLG + + D I+IGAFH+++ G++VV
Sbjct: 809 ---SCPGSAILAGFDDAIHDGVDILSLSLGGFGGTKTDLTTDPIAIGAFHSVQRGILVVC 979
Query: 295 GGGNSGPKFGTVTNVAPWLFSVAASTIDRNFVSYLQLGDKHIIMGTSLS-TGLPNEKFYS 353
GN G F TV N APW+ +VAASTIDR+ S + LG+ ++ G +++ + L N Y
Sbjct: 980 AAGNDGEPF-TVLNDAPWILTVAASTIDRDLQSDVVLGNNQVVKGRAINFSPLLNSPDYP 1156
Query: 354 LVSSVDAKVGN-ATIEDAKICKVGSLDPNKVKGKILFCLLRELDG---LVYAEEEAI--- 406
++ + A N + I DA+ C SLDP KV GKI+ C DG + Y+ +E I
Sbjct: 1157MIYAESAARANISNITDARQCHPDSLDPKKVIGKIVVC-----DGKNDIYYSTDEKIVIV 1321
Query: 407 -SGGSIGLVLGNDKQRGNDIMAYAHLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAKTE 465
+ G IGLV D Q G+ Y P + + G+ + YI +T P+ + T
Sbjct: 1322KALGGIGLVHITD-QSGSVAFYYVDF-PVTEVKSKHGDAILQYINSTSHPVGTILATVTI 1495
Query: 466 VGVKPAPVIASLSSRGPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIP--- 522
KPAP + SSRGP+ I +LKPDI APGV+IL A+ G +D +P
Sbjct: 1496PDYKPAPRVGYFSSRGPSLITSNVLKPDIAAPGVNILAAWFG--------NDTSEVPKGR 1651
Query: 523 ----YNIGSGTSISCPHVSAIVALLKTIYPNWSPAAFKSAIMTTTTIQGNNHRPIKDQSK 578
Y I SGTS++ PHVS + +K P WS +A KSAIMT+ N PI S
Sbjct: 1652KPSLYRILSGTSMATPHVSGLACSVKRKNPTWSASAIKSAIMTSAIQNDNLKGPITTDSG 1831
Query: 579 EDATPFGYGAGHIQPELAMDPGLVYDLNIVDYLNFLCAHGYNQTQMKMFS---RKPYICP 635
ATP+ YGAG I + PGLVY+ N VDYLN+LC +G N T +K+ S + + CP
Sbjct: 1832LIATPYDYGAGAITTSEPLQPGLVYETNNVDYLNYLCYNGLNITMIKVISGTVPENFNCP 2011
Query: 636 KSYN---MLDFNYPSITVPNLGKHFVQEVTRTVTNVGSPG--TYRVQVNEPHGIFVLIKP 690
K + + NYPSI V GK V+RTVTNV Y V P + V + P
Sbjct: 2012KDSSSDLISSINYPSIAVNFTGKADA-VVSRTVTNVDEEDETVYFPVVEAPSEVIVTLFP 2188
Query: 691 RSLTFNEVGEKKTFKIIFKVTKPTSSGYVFGHLLWSDGRHKVMSPLVV 738
+L F +K+++ I F+ K + +FG + WS+ ++ V P V+
Sbjct: 2189YNLEFTTSIKKQSYNITFR-PKTSLKKDLFGSITWSNDKYMVRIPFVL 2329
>TC226349 similar to PIR|JC7519|JC7519 subtilisin-like serine proteinase -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(56%)
Length = 1773
Score = 353 bits (906), Expect = 2e-97
Identities = 193/474 (40%), Positives = 288/474 (60%), Gaps = 7/474 (1%)
Frame = +1
Query: 272 EFFEDGISIGAFHAIENGVIVVAGGGNSGPKFGTVTNVAPWLFSVAASTIDRNFVSYLQL 331
+++ D ++IGAF A+E G++V GNSGP +++NVAPW+ +V A T+DR+F +Y+ L
Sbjct: 1 DYYRDSVAIGAFSAMEKGILVSCSAGNSGPGPYSLSNVAPWITTVGAGTLDRDFPAYVAL 180
Query: 332 GDKHIIMGTSLSTG--LPNEKFYSLVSSVDAKVGNATIEDAKICKVGSLDPNKVKGKILF 389
G+ G SL G LP+ SL V N + + +C G+L P KV GKI+
Sbjct: 181 GNGLNFSGVSLYRGNALPDS---SLPLVYAGNVSNGAM-NGNLCITGTLSPEKVAGKIVL 348
Query: 390 CLLRELDGLVYAEEEAISGGSIGLVLGNDKQRGNDIMAYAHLLPTSHINYTDGEYVHSYI 449
C R L V S G++G+VL N G +++A AHLLP + + G+ + Y+
Sbjct: 349 CD-RGLTARVQKGSVVKSAGALGMVLSNTAANGEELVADAHLLPATAVGQKAGDAIKKYL 525
Query: 450 KATKTPMAYMTKAKTEVGVKPAPVIASLSSRGPNPIQPIILKPDITAPGVDILYAYIGAI 509
+ P + T+VG++P+PV+A+ SSRGPN I P ILKPD+ APGV+IL + A+
Sbjct: 526 VSDAKPTVKIFFEGTKVGIQPSPVVAAFSSRGPNSITPQILKPDLIAPGVNILAGWSKAV 705
Query: 510 SPTGLASDNQWIPYNIGSGTSISCPHVSAIVALLKTIYPNWSPAAFKSAIMTTT-TIQGN 568
PTGL DN+ + +NI SGTS+SCPHVS + AL+K+ +P+WSPAA +SA+MTT T+
Sbjct: 706 GPTGLPVDNRRVDFNIISGTSMSCPHVSGLAALIKSAHPDWSPAAVRSALMTTAYTVYKT 885
Query: 569 NHRPIKDQSKEDATPFGYGAGHIQPELAMDPGLVYDLNIVDYLNFLCAHGYNQTQMKMFS 628
+ + + +TPF +G+GH+ P A++PGLVYDL + DYL FLCA Y+ ++ +
Sbjct: 886 GEKLQDSATGKPSTPFDHGSGHVDPVAALNPGLVYDLTVDDYLGFLCALNYSAAEISTLA 1065
Query: 629 RKPYICP--KSYNMLDFNYPSITVPNLGKHFVQEVTRTVTNVGSPGTYRVQV-NEPHGIF 685
++ + C K Y++ D NYPS V V + TRT+TNVG GTY+ V ++ +
Sbjct: 1066KRKFQCDAGKQYSVTDLNYPSFAVLFESSGSVVKHTRTLTNVGPAGTYKASVTSDTASVK 1245
Query: 686 VLIKPRSLTFNEVGEKKTFKIIFKVT-KPTSSGYVFGHLLWSDGRHKVMSPLVV 738
+ ++P+ L+F E EKKTF + F + P + FG + WSDG+H V SP+ V
Sbjct: 1246ISVEPQVLSFKE-NEKKTFTVTFSSSGSPQHTENAFGRVEWSDGKHLVGSPISV 1404
>TC229906 weakly similar to UP|Q9ZTT3 (Q9ZTT3) Subtilisin-like protease C1,
partial (40%)
Length = 1392
Score = 269 bits (688), Expect = 3e-72
Identities = 160/446 (35%), Positives = 243/446 (53%), Gaps = 6/446 (1%)
Frame = +2
Query: 299 SGPKFGTVTNVAPWLFSVAASTIDRNFVSYLQLGDKHIIMGTSLSTGLPNEKFYSLVSSV 358
SGP ++T +PW+ SVAASTI R F++ +QLG+ + G S++T K + LV +
Sbjct: 2 SGPGLSSITTYSPWILSVAASTIGRKFLTKVQLGNGMVFEGVSINTFDLKNKMFPLVYAG 181
Query: 359 DAK--VGNATIEDAKICKVGSLDPNKVKGKILFCLLRELDGLVYAEEEAISGGSIGLVLG 416
D ++ C V S+D + VKGKI+ C DG ++ G+ G++LG
Sbjct: 182 DVPNTADGYNSSTSRFCYVNSVDKHLVKGKIVLC-----DGNASPKKVGDLSGAAGMLLG 346
Query: 417 NDKQRGNDIMAYAHLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAKTEVGVKPAPVIAS 476
+ + + LPT+ I+ + + +HSY+ + + A + ++ + P I S
Sbjct: 347 ATDVKD---APFTYALPTAFISLRNFKLIHSYMVSLRNSTATIFRSDEDNDDSQTPFIVS 517
Query: 477 LSSRGPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISCPHV 536
SSRGPNP+ P LKPD+ APGV+IL A+ + + D + + YNI SGTS++CPHV
Sbjct: 518 FSSRGPNPLTPNTLKPDLAAPGVNILAAWSPVYTISEFKGDKRAVQYNIESGTSMACPHV 697
Query: 537 SAIVALLKTIYPNWSPAAFKSAIMTTTTIQGNNHRPIKDQSKEDATPFGYGAGHIQPELA 596
SA A +K+ +PNWSPA KSA+MTT T P+ DA F YGAG I P A
Sbjct: 698 SAAAAYVKSFHPNWSPAMIKSALMTTAT-------PMSPTLNPDA-EFAYGAGLINPLKA 853
Query: 597 MDPGLVYDLNIVDYLNFLCAHGYNQTQMKMFSRKPYICPKSYN---MLDFNYPSITVPNL 653
+PGLVYD++ DY+ FLC GY +++ ++ C K + D N PS+ +
Sbjct: 854 ANPGLVYDISEADYVKFLCGEGYTDEMLRVLTKDHSRCSKHAKKEAVYDLNLPSLALYVN 1033
Query: 654 GKHFVQEVTRTVTNVG-SPGTYRVQVNEPHGIFVLIKPRSLTFNEVGEKKTFKIIFKVTK 712
F + RTVTNVG + +Y+ +V P I + +KP L+F +G+KK+F +I +
Sbjct: 1034VSSFSRIFHRTVTNVGLATSSYKAKVVSPSLIDIQVKPNVLSFTSIGQKKSFSVI--IEG 1207
Query: 713 PTSSGYVFGHLLWSDGRHKVMSPLVV 738
+ + L+W DG +V SP+VV
Sbjct: 1208NVNPDILSASLVWDDGTFQVRSPIVV 1285
>TC226350 similar to PIR|JC7519|JC7519 subtilisin-like serine proteinase -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(34%)
Length = 1312
Score = 268 bits (685), Expect = 6e-72
Identities = 138/337 (40%), Positives = 213/337 (62%), Gaps = 5/337 (1%)
Frame = +1
Query: 407 SGGSIGLVLGNDKQRGNDIMAYAHLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAKTEV 466
S G++G+VL N G +++A AHLLP + + G+ + Y+ + P + T++
Sbjct: 7 SAGALGMVLSNTAANGEELVADAHLLPATAVGQKAGDAIKKYLFSDAKPTVKILFEGTKL 186
Query: 467 GVKPAPVIASLSSRGPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNIG 526
G++P+PV+A+ SSRGPN I P ILKPD+ APGV+IL + A+ PTGL DN+ + +NI
Sbjct: 187 GIQPSPVVAAFSSRGPNSITPQILKPDLIAPGVNILAGWSKAVGPTGLPVDNRRVDFNII 366
Query: 527 SGTSISCPHVSAIVALLKTIYPNWSPAAFKSAIMTTT-TIQGNNHRPIKDQSKEDATPFG 585
SGTS+SCPHVS + AL+K+ +P+WSPAA +SA+MTT T+ + + + +TPF
Sbjct: 367 SGTSMSCPHVSGLAALIKSAHPDWSPAAVRSALMTTAYTVYKTGEKLQDSATGKPSTPFD 546
Query: 586 YGAGHIQPELAMDPGLVYDLNIVDYLNFLCAHGYNQTQMKMFSRKPYICP--KSYNMLDF 643
+G+GH+ P A++PGLVYDL + DYL FLCA Y+ +++ +++ + C K Y++ D
Sbjct: 547 HGSGHVDPVAALNPGLVYDLTVDDYLGFLCALNYSASEINTLAKRKFQCDAGKQYSVTDL 726
Query: 644 NYPSITVPNLGKHFVQEVTRTVTNVGSPGTYRVQV-NEPHGIFVLIKPRSLTFNEVGEKK 702
NYPS V V+ TRT+TNVG GTY+ V ++ + + ++P+ L+F E EKK
Sbjct: 727 NYPSFAVLFESGGVVKH-TRTLTNVGPAGTYKASVTSDMASVKISVEPQVLSFKE-NEKK 900
Query: 703 TFKIIFKVT-KPTSSGYVFGHLLWSDGRHKVMSPLVV 738
+F + F + P FG + WSDG+H V +P+ +
Sbjct: 901 SFTVTFSSSGSPQQRVNAFGRVEWSDGKHVVGTPISI 1011
>TC207702 weakly similar to UP|Q8S896 (Q8S896) Subtilisin-like serine
protease AIR3 (Fragment), partial (20%)
Length = 1025
Score = 265 bits (678), Expect = 4e-71
Identities = 135/255 (52%), Positives = 178/255 (68%), Gaps = 4/255 (1%)
Frame = +2
Query: 489 ILKPDITAPGVDILYAYIGAISPTGLASDNQ-WIPYNIGSGTSISCPHVSAIVALLKTIY 547
ILKPD+TAPGV+IL AY S + L DN+ +N+ GTS+SCPHV+ I L+KT++
Sbjct: 5 ILKPDVTAPGVNILAAYSELASASNLLVDNRRGFKFNVLQGTSVSCPHVAGIAGLIKTLH 184
Query: 548 PNWSPAAFKSAIMTTTTIQGNNHRPIKDQ-SKEDATPFGYGAGHIQPELAMDPGLVYDLN 606
PNWSPAA KSAIMTT T N +RPI+D + A F YG+GH+QP+LA+DPGLVYDL+
Sbjct: 185 PNWSPAAIKSAIMTTATTLDNTNRPIQDAFDNKVADAFAYGSGHVQPDLAIDPGLVYDLS 364
Query: 607 IVDYLNFLCAHGYNQTQMKMFS-RKPYICPKSYNMLDFNYPSITVPNLGKHFVQEVTRTV 665
+ DYLNFLCA GY+Q + + +IC S+++ D NYPSIT+PNLG V +TRTV
Sbjct: 365 LADYLNFLCASGYDQQLISALNFNGTFICKGSHSVTDLNYPSITLPNLGLKPV-TITRTV 541
Query: 666 TNVGSPGTYRVQVNEPHGIFVLIKPRSLTFNEVGEKKTFKIIFKVTKPTS-SGYVFGHLL 724
TNVG P TY V+ P G +++ PRSLTF ++GEKK F++I + + T+ Y FG L
Sbjct: 542 TNVGPPATYTANVHSPAGYTIVVVPRSLTFTKIGEKKKFQVIVQASSVTTRRKYQFGDLR 721
Query: 725 WSDGRHKVMSPLVVK 739
W+DG+H V SP+ VK
Sbjct: 722 WTDGKHIVRSPITVK 766
>TC207663 similar to UP|P93204 (P93204) SBT1 protein (Subtilisin-like
protease) , partial (37%)
Length = 1171
Score = 262 bits (670), Expect = 4e-70
Identities = 136/304 (44%), Positives = 189/304 (61%), Gaps = 14/304 (4%)
Frame = +1
Query: 447 SYIKATKTPMAYMTKAKTEVGVKPAPVIASLSSRGPNPIQPIILKPDITAPGVDILYAYI 506
+Y+ ++ P A + T + V+P+PV+A+ SSRGPN + P ILKPD+ APGV+IL +
Sbjct: 7 NYVSSSPNPTAKIAFLGTHLQVQPSPVVAAFSSRGPNALTPKILKPDLIAPGVNILAGWT 186
Query: 507 GAISPTGLASDNQWIPYNIGSGTSISCPHVSAIVALLKTIYPNWSPAAFKSAIMTTTTIQ 566
GA+ PTGL D + + +NI SGTS+SCPHVS + A+LK +P WSPAA +SA+MTT
Sbjct: 187 GAVGPTGLTVDTRHVSFNIISGTSMSCPHVSGLAAILKGAHPQWSPAAIRSALMTTAYTS 366
Query: 567 GNNHRPIKDQSK-EDATPFGYGAGHIQPELAMDPGLVYDLNIVDYLNFLCAHGYNQTQMK 625
N I+D S + TPF YGAGH+ P A+DPGLVYD N+ DYL F CA Y+ Q+K
Sbjct: 367 YKNGETIQDISTGQPGTPFDYGAGHVDPVAALDPGLVYDANVDDYLGFFCALNYSSFQIK 546
Query: 626 MFSRKPYIC--PKSYNMLDFNYPSITVP-------NLGKHFVQEV--TRTVTNVGSPGTY 674
+ +R+ Y C K Y + DFNYPS VP G ++ V +R +TNVG+PGTY
Sbjct: 547 LAARRDYTCDPKKDYRVEDFNYPSFAVPMDTASGIGGGSDTLKTVKYSRVLTNVGAPGTY 726
Query: 675 RVQVNE--PHGIFVLIKPRSLTFNEVGEKKTFKIIFKVTKPTSSGYVFGHLLWSDGRHKV 732
+ V + +++P +L+F E+ EKK + + F T S F L W+DG+HKV
Sbjct: 727 KASVMSLGDSNVKTVVEPNTLSFTELYEKKDYTVSFTYTSMPSGTTSFARLEWTDGKHKV 906
Query: 733 MSPL 736
SP+
Sbjct: 907 GSPI 918
>TC229882 weakly similar to UP|Q75I27 (Q75I27) Putaive subtilisin-like
proteinase, partial (27%)
Length = 1263
Score = 235 bits (600), Expect = 5e-62
Identities = 126/326 (38%), Positives = 194/326 (58%), Gaps = 6/326 (1%)
Frame = +1
Query: 417 NDKQRGNDIMAYAHLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAKTEVGVKPAPVIAS 476
N G +++A +HLLP + G+ + +Y + P ++ T + VKP+PV+A+
Sbjct: 4 NTAASGEELVADSHLLPAVAVGRIVGDQIRAYASSDPNPTVHLDFRGTVLNVKPSPVVAA 183
Query: 477 LSSRGPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISCPHV 536
SSRGPN + ILKPD+ PGV+IL + AI P+GL+ D + +NI SGTS+SCPH+
Sbjct: 184 FSSRGPNMVTRQILKPDVIGPGVNILAGWSEAIGPSGLSDDTRKTQFNIMSGTSMSCPHI 363
Query: 537 SAIVALLKTIYPNWSPAAFKSAIMTTTTIQGNNHRPIKDQS-KEDATPFGYGAGHIQPEL 595
S + ALLK +P WS +A KSA+MTT + N ++D + + P+ +GAGH+ P
Sbjct: 364 SGLAALLKAAHPQWSSSAIKSALMTTADVHDNTKSQLRDAAGGAFSNPWAHGAGHVNPHK 543
Query: 596 AMDPGLVYDLNIVDYLNFLCAHGYNQTQMKMFSRKPYI-CPKSY-NMLDFNYPSITVPNL 653
A+ PGLVYD DY+ FLC+ Y ++++ +++ + C K + + NYPS +V
Sbjct: 544 ALSPGLVYDATPSDYIKFLCSLEYTPERIQLITKRSGVNCTKRFSDPGQLNYPSFSVLFG 723
Query: 654 GKHFVQEVTRTVTNVGSPGT-YRVQVNEPHGIFVLIKPRSLTFNEVGEKKTFKIIF--KV 710
GK V+ TR +TNVG G+ Y V V+ P + V +KP +L F +VGE++ + F K
Sbjct: 724 GKRVVR-YTRVLTNVGEAGSVYNVTVDAPSTVTVTVKPAALVFGKVGERQRYTATFVSKN 900
Query: 711 TKPTSSGYVFGHLLWSDGRHKVMSPL 736
S Y FG ++WS+ +H+V SP+
Sbjct: 901 GVGDSVRYGFGSIMWSNAQHQVRSPV 978
>TC221491 similar to UP|P93204 (P93204) SBT1 protein (Subtilisin-like
protease) , partial (34%)
Length = 872
Score = 234 bits (598), Expect = 8e-62
Identities = 119/276 (43%), Positives = 173/276 (62%), Gaps = 3/276 (1%)
Frame = +3
Query: 364 NATIEDAKICKVGSLDPNKVKGKILFCLLRELDGLVYAEEEAI--SGGSIGLVLGNDKQR 421
N + E +C G+L KV GKI+ C + G E+ + S G IG++L N++
Sbjct: 54 NVSDESQNLCTRGTLIAEKVAGKIVIC---DRGGNARVEKGLVVKSAGGIGMILSNNEDY 224
Query: 422 GNDIMAYAHLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAKTEVGVKPAPVIASLSSRG 481
G +++A ++LLP + + + Y+ ++ P A + T++GV+P+PV+A+ SSRG
Sbjct: 225 GEELVADSYLLPAAALGQKSSNELKKYVFSSPNPTAKLGFGGTQLGVQPSPVVAAFSSRG 404
Query: 482 PNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISCPHVSAIVA 541
PN + P ILKPD+ APGV+IL + GA+ PTGL D + + +NI SGTS+SCPHV+ + A
Sbjct: 405 PNVLTPKILKPDLIAPGVNILAGWTGAVGPTGLTEDTRHVEFNIISGTSMSCPHVTGLAA 584
Query: 542 LLKTIYPNWSPAAFKSAIMTTTTIQGNNHRPIKDQSKE-DATPFGYGAGHIQPELAMDPG 600
LLK +P WSPAA +SA+MTT N + IKD + ATPF YGAGH+ P A DPG
Sbjct: 585 LLKGTHPEWSPAAIRSALMTTAYRTYKNGQTIKDVATGLPATPFDYGAGHVDPVAAFDPG 764
Query: 601 LVYDLNIVDYLNFLCAHGYNQTQMKMFSRKPYICPK 636
LVYD + DYL+F CA Y+ Q+K+ +R+ + C K
Sbjct: 765 LVYDTTVDDYLSFFCALNYSPYQIKLVARRDFTCSK 872
>TC226351 similar to PIR|JC7519|JC7519 subtilisin-like serine proteinase -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(48%)
Length = 1490
Score = 212 bits (540), Expect(2) = 1e-61
Identities = 123/291 (42%), Positives = 179/291 (61%), Gaps = 8/291 (2%)
Frame = +3
Query: 135 SDDGMGPVPSRWRGICQ----LDNFHCNRKLIGARFYSQGYESKFGRLNQS--LYNARDV 188
++ G+GPVPS W+G C+ L +CNRKLIGARF+S+G E+ G +N++ +ARD
Sbjct: 648 TNTGLGPVPSTWKGACETGTNLTASNCNRKLIGARFFSKGVEAILGPINETEESRSARDD 827
Query: 189 LGHGTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVCWLGTI*IECTDADIMQ 248
GHGT T S A G+ VS A++FG A+GTA+G + R+ VAAYKVCW G C +DI+
Sbjct: 828 DGHGTHTASTAAGSVVSDASLFGYASGTARGMATRARVAAYKVCWKGG----CFSSDILA 995
Query: 249 AFEDAISDGVDIISCSLGQTSPKEFFEDGISIGAFHAIENGVIVVAGGGNSGPKFGTVTN 308
A E AI D V+++S SLG +++ D ++IGAF A+ENG++V GN+GP +++N
Sbjct: 996 AIERAILDNVNVLSLSLGG-GMSDYYRDSVAIGAFSAMENGILVSCSAGNAGPSPYSLSN 1172
Query: 309 VAPWLFSVAASTIDRNFVSYLQLGDKHIIMGTSLSTG--LPNEKFYSLVSSVDAKVGNAT 366
VAPW+ +V A T+DR+F +Y+ LG+ G SL G +P+ + + V N
Sbjct: 1173VAPWITTVGAGTLDRDFPAYVALGNGLNFSGVSLYRGNAVPDSPLPFVYA---GNVSNGA 1343
Query: 367 IEDAKICKVGSLDPNKVKGKILFCLLRELDGLVYAEEEAISGGSIGLVLGN 417
+ + +C G+L P KV GKI+ C R L V S G++G+VL N
Sbjct: 1344M-NGNLCITGTLSPEKVAGKIVLC-DRGLTARVQKGSVVKSAGALGMVLSN 1490
Score = 43.9 bits (102), Expect(2) = 1e-61
Identities = 29/105 (27%), Positives = 52/105 (48%)
Frame = +2
Query: 1 SYIVYIGSHSHGPNPSASDLQSATDSHYNLLGSHLGSHEKAKEAIFYSYNKHINGFAAVL 60
+YIV++ S++ + + H S L + + E I Y+Y+ I+G+A L
Sbjct: 290 TYIVHVAK---------SEMPESFEHHALWYESSLKTVSDSAE-IMYTYDNAIHGYATRL 439
Query: 61 EVEEAAKIAKHPNVVSVFENKGHELQTTRSWEFLGLENNYGVVPK 105
EEA + +++V +EL TTR+ FLGL+ + + P+
Sbjct: 440 TAEEARVLETQAGILAVLPETRYELHTTRTPMFLGLDKSANMFPE 574
>TC233666 weakly similar to UP|Q9FJF3 (Q9FJF3) Serine protease-like protein,
partial (30%)
Length = 678
Score = 223 bits (568), Expect = 2e-58
Identities = 115/227 (50%), Positives = 150/227 (65%)
Frame = +1
Query: 192 GTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVCWLGTI*IECTDADIMQAFE 251
G+ TLS GG FV GANVFGL NGTA+GGSPR+ VA YKVCW EC DADIM AF+
Sbjct: 4 GSHTLSTIGGTFVPGANVFGLGNGTAEGGSPRARVATYKVCWPPIDGNECFDADIMAAFD 183
Query: 252 DAISDGVDIISCSLGQTSPKEFFEDGISIGAFHAIENGVIVVAGGGNSGPKFGTVTNVAP 311
AI DGVD++S SLG + ++F+DG+SIGAFHA G+ V+ GN GP TV NVAP
Sbjct: 184 MAIHDGVDVLSLSLGGNAT-DYFDDGLSIGAFHANMKGIPVICSAGNYGPTPATVFNVAP 360
Query: 312 WLFSVAASTIDRNFVSYLQLGDKHIIMGTSLSTGLPNEKFYSLVSSVDAKVGNATIEDAK 371
W+ +V AST+DR F S ++L + MG SLS +P +K Y L+++ DAK N +E+A
Sbjct: 361 WILTVGASTLDRQFDSVVELHNGQRFMGASLSKAMPEDKLYPLINAADAKAANKPVENAT 540
Query: 372 ICKVGSLDPNKVKGKILFCLLRELDGLVYAEEEAISGGSIGLVLGND 418
+C G++DP K +GKIL C LR + V A+ G+ ++L ND
Sbjct: 541 LCMRGTIDPEKARGKILVC-LRGVTARVEKSLVALEAGAAXMILCND 678
>TC228967 similar to GB|BAB09208.1|9758669|AB012245 subtilisin-like protease
{Arabidopsis thaliana;} , partial (21%)
Length = 1106
Score = 220 bits (561), Expect = 2e-57
Identities = 127/299 (42%), Positives = 163/299 (54%), Gaps = 9/299 (3%)
Frame = +2
Query: 449 IKATKTPMAYMTKAKTEVGVKPAPVIASLSSRGPNPIQPIILKPDITAPGVDILYAYIGA 508
+ +T PMA + T + KPAP +AS SSRGPN + P ILKPDITAPGVDIL A+
Sbjct: 5 VHSTPNPMAQILPGTTVLETKPAPSMASFSSRGPNIVDPNILKPDITAPGVDILAAWTAE 184
Query: 509 ISPTGLA-SDNQWIPYNIGSGTSISCPHVSAIVALLKTIYPNWSPAAFKSAIMTTTTIQG 567
PT + +D + + YNI SGTS+SCPHV+A LLK I+P WS AA +SA+MTT
Sbjct: 185 DGPTRMTFNDKRVVKYNIFSGTSMSCPHVAAAAVLLKAIHPTWSTAAIRSALMTTAMTTD 364
Query: 568 NNHRPIKDQSKEDATPFGYGAGHIQPELAMDPGLVYDLNIVDYLNFLCAHGYNQTQMKMF 627
N P+ D++ ATPF G+GH P+ A DPGLVYD + + YL + C G Q
Sbjct: 365 NTGHPLTDETGNPATPFAMGSGHFNPKRAADPGLVYDASYMGYLLYTCNLGVTQN----- 529
Query: 628 SRKPYICPKSY-NMLDFNYPSITVPNLGKHFVQEVTRTVTNVG-SPGTYRVQVNEPHGIF 685
Y CPKS+ + NYPSI + L ++ + + RTVTNVG Y+ P
Sbjct: 530 FNITYNCPKSFLEPFELNYPSIQIHRL--YYTKTIKRTVTNVGRGRSVYKFSAVSPKEYS 703
Query: 686 VLIKPRSLTFNEVGEKKTFKIIFKV------TKPTSSGYVFGHLLWSDGRHKVMSPLVV 738
+ P L FN VG+K F I TK Y FG W+ H V SP+ V
Sbjct: 704 ITATPNILKFNHVGQKINFTITVTANWSQIPTKHGPDKYYFGWYAWTHQHHIVRSPVAV 880
>TC216428 similar to UP|Q9AX30 (Q9AX30) Subtilisin-like protease, partial
(12%)
Length = 810
Score = 220 bits (560), Expect = 2e-57
Identities = 110/197 (55%), Positives = 137/197 (68%), Gaps = 2/197 (1%)
Frame = +1
Query: 544 KTIYPNWSPAAFKSAIMTTTTIQGNNHRPIKDQSKEDATPFGYGAGHIQPELAMDPGLVY 603
KT +P WSPAA KSAIMTT T N ++PI++ + ATPF YGAGHIQP LA+DPGLVY
Sbjct: 1 KTYHPTWSPAAIKSAIMTTATTLDNTNQPIRNAFHKVATPFEYGAGHIQPNLAIDPGLVY 180
Query: 604 DLNIVDYLNFLCAHGYNQTQMKMFSRK--PYICPKSYNMLDFNYPSITVPNLGKHFVQEV 661
DL DYLNFLCA GYNQ + +F++ PY CPKSY + DFNYPSITV + G + V
Sbjct: 181 DLRTTDYLNFLCASGYNQALLNLFAKLKFPYTCPKSYRIEDFNYPSITVRHPGSKTI-SV 357
Query: 662 TRTVTNVGSPGTYRVQVNEPHGIFVLIKPRSLTFNEVGEKKTFKIIFKVTKPTSSGYVFG 721
TRTVTNVG P TY V + P GI VL++P SLTF GEKK F++I + +FG
Sbjct: 358 TRTVTNVGPPSTYVVNTHGPKGIKVLVQPSSLTFKRTGEKKKFQVILQPIGARRG--LFG 531
Query: 722 HLLWSDGRHKVMSPLVV 738
+L W+DG+H+V SP+ +
Sbjct: 532 NLSWTDGKHRVTSPITI 582
>TC229044 similar to UP|Q93WQ0 (Q93WQ0) Subtilisin-type protease, partial
(42%)
Length = 984
Score = 219 bits (558), Expect = 3e-57
Identities = 129/314 (41%), Positives = 180/314 (57%), Gaps = 8/314 (2%)
Frame = +1
Query: 433 PTSHINYTDGEYVHSYIKATKTPMAYMTKAKTEVGVKPAPVIASLSSRGPNPIQPIILKP 492
P + I+ DG + YI +T P+A + T + KPAPV+ + SSRGP+ + ILKP
Sbjct: 16 PATVISSKDGVTILQYINSTSNPVATILPTATVLDYKPAPVVPNFSSRGPSSLSSNILKP 195
Query: 493 DITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISCPHVSAIVALLKTIYPNWSP 552
DI APGV+IL A+IG + + + YNI SGTS++CPHVS + + +KT P WS
Sbjct: 196 DIAAPGVNILAAWIGN-NADDVPKGRKPSLYNIISGTSMACPHVSGLASSVKTRNPTWSA 372
Query: 553 AAFKSAIMTTTTIQGNNHRPIKDQSKEDATPFGYGAGHIQPELAMDPGLVYDLNIVDYLN 612
+A KSAIMT+ N PI S ATP+ YGAG + ++ PGLVY+ N +DYLN
Sbjct: 373 SAIKSAIMTSAIQINNLKAPITTDSGRVATPYDYGAGEMTTSESLQPGLVYETNTIDYLN 552
Query: 613 FLCAHGYNQTQMKMFSR---KPYICPKSYN---MLDFNYPSITVPNLGKHFVQEVTRTVT 666
+LC G N T +K+ SR + CPK + + + NYPSI V GK V V+RTVT
Sbjct: 553 YLCYIGLNITTVKVISRTVPANFSCPKDSSSDLISNINYPSIAVNFTGKAAV-NVSRTVT 729
Query: 667 NVGSPG--TYRVQVNEPHGIFVLIKPRSLTFNEVGEKKTFKIIFKVTKPTSSGYVFGHLL 724
NVG Y V P G+ V + P L F + +K +++IF T + +FG +
Sbjct: 730 NVGEEDETAYSPVVEAPSGVKVTVTPDKLQFTKSSKKLGYQVIFSSTLTSLKEDLFGSIT 909
Query: 725 WSDGRHKVMSPLVV 738
WS+G++ V SP V+
Sbjct: 910 WSNGKYMVRSPFVL 951
>TC216758 similar to PIR|T05768|T05768 subtilisin-like proteinase -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(35%)
Length = 796
Score = 216 bits (551), Expect = 2e-56
Identities = 123/274 (44%), Positives = 162/274 (58%), Gaps = 5/274 (1%)
Frame = +3
Query: 289 GVIVVAGGGNSGPKFGTVTNVAPWLFSVAASTIDRNFVSYLQLGDKHIIMGTSLSTGLPN 348
GV V + GN GP +VTN+APWL +V A TIDR+F S + LGD + G SL G
Sbjct: 3 GVFVSSSAGNDGPSGMSVTNLAPWLTTVGAGTIDRDFPSQVILGDGRRLSGVSLYAGAAL 182
Query: 349 E-KFYSLVSSVDAKVGNATIEDAKICKVGSLDPNKVKGKILFC----LLRELDGLVYAEE 403
+ K Y LV G + I +C SLDPN VKGKI+ C R GLV +
Sbjct: 183 KGKMYQLVYP-----GKSGILGDSLCMENSLDPNMVKGKIVICDRGSSPRVAKGLVVKK- 344
Query: 404 EAISGGSIGLVLGNDKQRGNDIMAYAHLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAK 463
G +G++L N G ++ AHLLP + +G+ + YI ++ P A +
Sbjct: 345 ----AGGVGMILANGISNGEGLVGDAHLLPACAVGANEGDVIKKYISSSTNPTATLDFKG 512
Query: 464 TEVGVKPAPVIASLSSRGPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPY 523
T +G+KPAPVIAS S+RGPN + P ILKPD APGV+IL A+ A+ PTGL SD + +
Sbjct: 513 TILGIKPAPVIASFSARGPNGLNPQILKPDFIAPGVNILAAWTQAVGPTGLDSDTRRTEF 692
Query: 524 NIGSGTSISCPHVSAIVALLKTIYPNWSPAAFKS 557
NI SGTS++CPHVS ALLK+ +P+WSPAA +S
Sbjct: 693 NILSGTSMACPHVSGAAALLKSAHPDWSPAALRS 794
>TC228968 similar to GB|BAB09208.1|9758669|AB012245 subtilisin-like protease
{Arabidopsis thaliana;} , partial (20%)
Length = 1008
Score = 205 bits (522), Expect = 5e-53
Identities = 115/302 (38%), Positives = 175/302 (57%), Gaps = 8/302 (2%)
Frame = +3
Query: 198 VAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVCWL--------GTI*IECTDADIMQA 249
VAG + + + G A GTA GG+P + +A YK CW G I CT+ D+++A
Sbjct: 6 VAGRVVPNASAIGGFAKGTALGGAPLARLAIYKACWPIKGKSKHEGNI---CTNIDMLKA 176
Query: 250 FEDAISDGVDIISCSLGQTSPKEFFEDGISIGAFHAIENGVIVVAGGGNSGPKFGTVTNV 309
+DAI DGVD++S S+G ++P + ED I+ GA HA+ ++VV GNSGP T++N
Sbjct: 177 IDDAIGDGVDVLSISIGFSAPISYEEDVIARGALHAVRKNIVVVCSAGNSGPLPQTLSNP 356
Query: 310 APWLFSVAASTIDRNFVSYLQLGDKHIIMGTSLSTGLPNEKFYSLVSSVDAKVGNATIED 369
APW+ +VAAST+DR+F + ++L II G S++ FY LV + D + +
Sbjct: 357 APWIITVAASTVDRSFHAPIKLNGT-IIEGRSITPLHMGNSFYPLVLARDVEHPGLPSNN 533
Query: 370 AKICKVGSLDPNKVKGKILFCLLRELDGLVYAEEEAISGGSIGLVLGNDKQRGNDIMAYA 429
+ C +L PNK +GKI+ C+ + + L E +GG +G +LGN+K G D+ +
Sbjct: 534 SGFCLDNTLQPNKARGKIVLCMRGQGERLKKGLEVQRAGG-VGFILGNNKLNGKDVPSDP 710
Query: 430 HLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAKTEVGVKPAPVIASLSSRGPNPIQPII 489
H +P + ++Y + + Y+ +T PMA + T + KPAP +AS SSRGPN + P I
Sbjct: 711 HFIPATGVSYENSLKLIQYVHSTPNPMAQILPGTTVLETKPAPSMASFSSRGPNIVDPNI 890
Query: 490 LK 491
LK
Sbjct: 891 LK 896
>TC232495 similar to UP|Q8W554 (Q8W554) AT3g14240/MLN21_2, partial (40%)
Length = 731
Score = 203 bits (516), Expect = 3e-52
Identities = 102/233 (43%), Positives = 153/233 (64%), Gaps = 5/233 (2%)
Frame = +3
Query: 422 GNDIMAYAHLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAK-TEVGVKPAPVIASLSSR 480
G ++A H+LP + + T G+ + SYI ++TP K T +GV+PAPV+AS S+R
Sbjct: 9 GEGLVADCHVLPATAVGATAGDEIRSYIGNSRTPATATIVFKGTRLGVRPAPVVASFSAR 188
Query: 481 GPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISCPHVSAIV 540
GPNP+ P ILKPD+ APG++IL A+ + P+G+ SD + +NI SGTS++CPHVS +
Sbjct: 189 GPNPVSPEILKPDVIAPGLNILAAWPDHVGPSGVPSDGRRTEFNILSGTSMACPHVSGLA 368
Query: 541 ALLKTIYPNWSPAAFKSAIMTTTTIQGNNHRPIKDQSKED-ATPFGYGAGHIQPELAMDP 599
ALLK +P+WSPA+ +SA+MTT N PI D+S + ++ F YGAGH+ P AM+P
Sbjct: 369 ALLKAAHPDWSPASIRSALMTTAYTVDNKGDPILDESTGNVSSVFDYGAGHVHPVKAMNP 548
Query: 600 GLVYDLNIVDYLNFLCAHGYNQTQMKMFSRKPYIC---PKSYNMLDFNYPSIT 649
GLVYD++ DY+NFLC Y +++ +R+ C ++ + + NYPS++
Sbjct: 549 GLVYDISSNDYVNFLCNSNYTTNTIRVITRRNADCSGAKRAGHSGNLNYPSLS 707
>TC222231 weakly similar to UP|Q9FVC7 (Q9FVC7) Subtilase , partial (17%)
Length = 807
Score = 184 bits (468), Expect = 9e-47
Identities = 97/219 (44%), Positives = 136/219 (61%), Gaps = 3/219 (1%)
Frame = +1
Query: 524 NIGSGTSISCPHVSAIVALLKTIYPNWSPAAFKSAIMTTTTIQGNNHRPI-KDQSKEDAT 582
NI SGTS++CP+V+ I L+K ++P+WSP+A KSAIMTT T+ +HRPI D + A
Sbjct: 1 NILSGTSMACPNVTGIATLVKAVHPSWSPSAIKSAIMTTATVLDKHHRPITADPEQRRAN 180
Query: 583 PFGYGAGHIQPELAMDPGLVYDLNIVDYLNFLCAHGYNQTQMKMFSRKPYICPKSYNML- 641
F YG+G + P +DPGL+YD D++ FLC+ GY+Q + +R C ++++
Sbjct: 181 AFDYGSGFVNPARVLDPGLIYDSKPADFVAFLCSLGYDQRSLHQVTRDNSTCDRAFSTAS 360
Query: 642 DFNYPSITVPNLGKHFVQEVTRTVTNVG-SPGTYRVQVNEPHGIFVLIKPRSLTFNEVGE 700
D NYPSI VPNL +F VTR VTNVG + Y+ V+ P G V + P L F +G+
Sbjct: 361 DLNYPSIAVPNLKDNF--SVTRIVTNVGKARSVYKAVVSSPPGXXVSVIPNRLIFTRIGQ 534
Query: 701 KKTFKIIFKVTKPTSSGYVFGHLLWSDGRHKVMSPLVVK 739
K F + FK++ P S GY FG L W + +V SPLVV+
Sbjct: 535 KINFTVNFKLSAP-SKGYAFGFLSWRNRISQVTSPLVVR 648
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.136 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,321,670
Number of Sequences: 63676
Number of extensions: 473694
Number of successful extensions: 2478
Number of sequences better than 10.0: 131
Number of HSP's better than 10.0 without gapping: 2286
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2306
length of query: 740
length of database: 12,639,632
effective HSP length: 104
effective length of query: 636
effective length of database: 6,017,328
effective search space: 3827020608
effective search space used: 3827020608
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC147712.15


