

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147712.11 + phase: 0
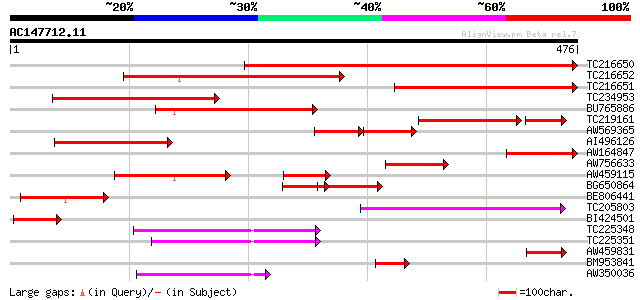
(476 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC216650 homologue to UP|Q9LG32 (Q9LG32) F14J16.5, partial (50%) 541 e-154
TC216652 similar to UP|Q9LG32 (Q9LG32) F14J16.5, partial (35%) 340 8e-94
TC216651 similar to GB|AAD28199.1|4704662|AF116860 uracil phosph... 286 1e-77
TC234953 similar to UP|Q9LFZ2 (Q9LFZ2) F20N2.19, partial (11%) 259 1e-69
BU765886 homologue to GP|10177966|db uridine kinase-like protein... 230 9e-61
TC219161 homologue to GB|AAM10488.1|29465725|AY089970 uracil pho... 155 7e-51
AW569365 homologue to GP|10177966|dbj uridine kinase-like protei... 93 4e-35
AI496126 similar to GP|10177966|dbj uridine kinase-like protein ... 135 3e-32
AW164847 114 1e-25
AW756633 homologue to GP|8778301|gb|A F14J16.5 {Arabidopsis thal... 111 8e-25
AW459115 weakly similar to GP|10177966|db uridine kinase-like pr... 102 5e-22
BG650864 97 2e-20
BE806441 similar to GP|9294212|dbj| uridine kinase-like protein ... 83 2e-16
TC205803 similar to UP|UPP_TOBAC (P93394) Uracil phosphoribosylt... 82 4e-16
BI424501 73 3e-13
TC225348 homologue to UP|KPPR_MESCR (P27774) Phosphoribulokinase... 70 3e-12
TC225351 homologue to UP|P93681 (P93681) Phosphoribulokinase (F... 64 2e-10
AW459831 61 1e-09
BM953841 similar to GP|7939517|dbj| uridine kinase-like protein ... 55 7e-08
AW350036 homologue to SP|P25697|KPPR Phosphoribulokinase chloro... 53 3e-07
>TC216650 homologue to UP|Q9LG32 (Q9LG32) F14J16.5, partial (50%)
Length = 1050
Score = 541 bits (1395), Expect = e-154
Identities = 268/280 (95%), Positives = 275/280 (97%), Gaps = 1/280 (0%)
Frame = +1
Query: 198 AILDQYSKFVKPAFDDFILPTKKYADIIIPRGGDNHVAVDLIVQHIRTKLGQHDLCKIYP 257
A+LDQYSKFVKPAFDDFILPTKKYADIIIPRGGDNHVA+DLIVQHIRTKLGQHDLCKIYP
Sbjct: 1 AVLDQYSKFVKPAFDDFILPTKKYADIIIPRGGDNHVAIDLIVQHIRTKLGQHDLCKIYP 180
Query: 258 NLYVIHSTFQIRGMHTLIRDAQITKHDFVFYSDRLIRLVVEHGLGHLPFTEKQVITPTGS 317
NLYVI STFQIRGMHTLIR A+ TKHDF+FYSDRLIRLVVEHGLGHLPFTEKQVITPTGS
Sbjct: 181 NLYVIQSTFQIRGMHTLIRHAKTTKHDFIFYSDRLIRLVVEHGLGHLPFTEKQVITPTGS 360
Query: 318 VYTGVDFCKRLCGVSVIRSGESMENALRACCKGIKIGKILIHREGDNGQQLIYEKLPNDI 377
VYTGVDFCKRLCGVSVIRSGESMENALRACCKGIKIGKILIHREGDNGQQLIYEKLPNDI
Sbjct: 361 VYTGVDFCKRLCGVSVIRSGESMENALRACCKGIKIGKILIHREGDNGQQLIYEKLPNDI 540
Query: 378 SERHVLLLDPILGTGNSAVQAISLLIQKGVPESNIIFLNLISAPKGVHVVCKCFPRIKIV 437
S+RHVLLLDPILGTGNSAVQAISLLI+KGVPESNIIFLNLISAPKGVHVVCK FPRIKIV
Sbjct: 541 SDRHVLLLDPILGTGNSAVQAISLLIKKGVPESNIIFLNLISAPKGVHVVCKSFPRIKIV 720
Query: 438 TSEIEIGLNEDFRVIPGMGEFGDRYFGT-DDDEQVVVASQ 476
TSEIEIGLNEDFRVIPGMGEFGDRYFGT DDDEQVVV+SQ
Sbjct: 721 TSEIEIGLNEDFRVIPGMGEFGDRYFGTDDDDEQVVVSSQ 840
>TC216652 similar to UP|Q9LG32 (Q9LG32) F14J16.5, partial (35%)
Length = 566
Score = 340 bits (872), Expect = 8e-94
Identities = 165/188 (87%), Positives = 177/188 (93%), Gaps = 2/188 (1%)
Frame = +3
Query: 96 RVQDYNFDHPDAFDTEELLRVMDKLKHGEAVDIPKYDFKSYKSDAL--RRVNPSDVIILE 153
RVQDYNFDHP+AFDTE+LLRVMDKLKH +AVDIPKYDFK YK+D RRVNP+DVIILE
Sbjct: 3 RVQDYNFDHPEAFDTEQLLRVMDKLKHSQAVDIPKYDFKGYKNDVFPARRVNPADVIILE 182
Query: 154 GILVFHDPRVRELMNMKIFVDTDADVRLARRIRRDTSEKDRDIGAILDQYSKFVKPAFDD 213
GILVFHDPRVR LMNMKIFVDTDADVRLARRI+RDT++ R+IGA+LDQYSKFVKPAFDD
Sbjct: 183 GILVFHDPRVRALMNMKIFVDTDADVRLARRIKRDTADNAREIGAVLDQYSKFVKPAFDD 362
Query: 214 FILPTKKYADIIIPRGGDNHVAVDLIVQHIRTKLGQHDLCKIYPNLYVIHSTFQIRGMHT 273
FILPTKKYADIIIPRGGDNHVA+DLIVQHIRTKLGQHDLCKIYPNLYVI STFQIRGMHT
Sbjct: 363 FILPTKKYADIIIPRGGDNHVAIDLIVQHIRTKLGQHDLCKIYPNLYVIQSTFQIRGMHT 542
Query: 274 LIRDAQIT 281
LIRD+Q T
Sbjct: 543 LIRDSQTT 566
>TC216651 similar to GB|AAD28199.1|4704662|AF116860 uracil
phosphoribosyltransferase 1 {Arabidopsis thaliana;} ,
partial (84%)
Length = 829
Score = 286 bits (733), Expect = 1e-77
Identities = 142/154 (92%), Positives = 150/154 (97%), Gaps = 1/154 (0%)
Frame = +2
Query: 324 FCKRLCGVSVIRSGESMENALRACCKGIKIGKILIHREGDNGQQLIYEKLPNDISERHVL 383
FCKRLCGVS+IRSGESMENALRACCKGIKIGKILIHREGDNGQQLIYEKLPNDIS+RHVL
Sbjct: 2 FCKRLCGVSIIRSGESMENALRACCKGIKIGKILIHREGDNGQQLIYEKLPNDISDRHVL 181
Query: 384 LLDPILGTGNSAVQAISLLIQKGVPESNIIFLNLISAPKGVHVVCKCFPRIKIVTSEIEI 443
LLDPILGTGNSAVQAISLL++KGVPESNIIFLNL+SAP+GVHVVCK FPRIKI+TSEIEI
Sbjct: 182 LLDPILGTGNSAVQAISLLLKKGVPESNIIFLNLVSAPQGVHVVCKRFPRIKILTSEIEI 361
Query: 444 GLNEDFRVIPGMGEFGDRYFGTD-DDEQVVVASQ 476
GLNEDFRVIPGMGEFGDRYFGTD DD+QVV SQ
Sbjct: 362 GLNEDFRVIPGMGEFGDRYFGTDNDDQQVVPPSQ 463
>TC234953 similar to UP|Q9LFZ2 (Q9LFZ2) F20N2.19, partial (11%)
Length = 497
Score = 259 bits (663), Expect = 1e-69
Identities = 128/141 (90%), Positives = 133/141 (93%), Gaps = 1/141 (0%)
Frame = +3
Query: 37 PTTSATDVYNQPFVIGVAGGAASGKKTVCDMIVQQLHDQRVVLVNQDSFYNNLTEEERAR 96
PTTS TD+Y QPFVIGVAGGAASGK TVCDMI+QQLHDQRVVLVNQDSFYNNLTEEE R
Sbjct: 3 PTTSETDMYKQPFVIGVAGGAASGKTTVCDMIIQQLHDQRVVLVNQDSFYNNLTEEELTR 182
Query: 97 VQDYNFDHPDAFDTEELLRVMDKLKHGEAVDIPKYDFKSYKS-DALRRVNPSDVIILEGI 155
VQDYNFDHPDAFDT++LL VMDKLKHGEAVDIPKYDFKSYKS D LRRVNPSDVIILEGI
Sbjct: 183 VQDYNFDHPDAFDTKQLLHVMDKLKHGEAVDIPKYDFKSYKSDDVLRRVNPSDVIILEGI 362
Query: 156 LVFHDPRVRELMNMKIFVDTD 176
LVFHDPRVRELMNMKIFVDT+
Sbjct: 363 LVFHDPRVRELMNMKIFVDTE 425
>BU765886 homologue to GP|10177966|db uridine kinase-like protein
{Arabidopsis thaliana}, partial (28%)
Length = 421
Score = 230 bits (587), Expect = 9e-61
Identities = 110/138 (79%), Positives = 126/138 (90%), Gaps = 2/138 (1%)
Frame = +3
Query: 123 GEAVDIPKYDFKSYK--SDALRRVNPSDVIILEGILVFHDPRVRELMNMKIFVDTDADVR 180
G+ V +P YDFK ++ SD+ R+VN SDVIILEGILVFHD RVR+LMNMKIFVDTDADVR
Sbjct: 3 GQGVHVPIYDFKKHQRSSDSFRQVNASDVIILEGILVFHDQRVRDLMNMKIFVDTDADVR 182
Query: 181 LARRIRRDTSEKDRDIGAILDQYSKFVKPAFDDFILPTKKYADIIIPRGGDNHVAVDLIV 240
LARRIRRDT E+ RDI ++L+QY+KFVKPAFDDF+LP+KKYAD+IIPRGGDNHVA+DLIV
Sbjct: 183 LARRIRRDTMERGRDINSVLEQYAKFVKPAFDDFVLPSKKYADVIIPRGGDNHVAIDLIV 362
Query: 241 QHIRTKLGQHDLCKIYPN 258
QHIRTKLGQHDLCKIYPN
Sbjct: 363 QHIRTKLGQHDLCKIYPN 416
>TC219161 homologue to GB|AAM10488.1|29465725|AY089970 uracil
phosphoribosyltransferase {Arabidopsis thaliana;} ,
partial (25%)
Length = 642
Score = 155 bits (393), Expect(2) = 7e-51
Identities = 75/86 (87%), Positives = 81/86 (93%)
Frame = +2
Query: 344 LRACCKGIKIGKILIHREGDNGQQLIYEKLPNDISERHVLLLDPILGTGNSAVQAISLLI 403
LRACCKGIKIGKILIHR+GDNG+QLIYEKLP DISERHVLLLDP+L TGNSA QAI LLI
Sbjct: 2 LRACCKGIKIGKILIHRDGDNGKQLIYEKLPKDISERHVLLLDPVLATGNSANQAIELLI 181
Query: 404 QKGVPESNIIFLNLISAPKGVHVVCK 429
QKGVPES+IIFLNLISAP+G+H VCK
Sbjct: 182 QKGVPESHIIFLNLISAPEGIHCVCK 259
Score = 63.5 bits (153), Expect(2) = 7e-51
Identities = 27/34 (79%), Positives = 33/34 (96%)
Frame = +1
Query: 434 IKIVTSEIEIGLNEDFRVIPGMGEFGDRYFGTDD 467
+KIVTSEI+I +NE++RVIPG+GEFGDRYFGTDD
Sbjct: 271 LKIVTSEIDIEINEEYRVIPGLGEFGDRYFGTDD 372
>AW569365 homologue to GP|10177966|dbj uridine kinase-like protein
{Arabidopsis thaliana}, partial (17%)
Length = 264
Score = 92.8 bits (229), Expect(2) = 4e-35
Identities = 40/44 (90%), Positives = 44/44 (99%)
Frame = +3
Query: 298 EHGLGHLPFTEKQVITPTGSVYTGVDFCKRLCGVSVIRSGESME 341
EHGLGHLPFTEKQV+TPTGSVYTGVDFCK+LCGVS++RSGESME
Sbjct: 132 EHGLGHLPFTEKQVVTPTGSVYTGVDFCKKLCGVSIVRSGESME 263
Score = 73.9 bits (180), Expect(2) = 4e-35
Identities = 35/41 (85%), Positives = 36/41 (87%)
Frame = +2
Query: 257 PNLYVIHSTFQIRGMHTLIRDAQITKHDFVFYSDRLIRLVV 297
PN YVI STFQIRGMHTLIRD I+KHDFVFYSDRLI LVV
Sbjct: 8 PNAYVIQSTFQIRGMHTLIRDRDISKHDFVFYSDRLIHLVV 130
>AI496126 similar to GP|10177966|dbj uridine kinase-like protein {Arabidopsis
thaliana}, partial (18%)
Length = 333
Score = 135 bits (341), Expect = 3e-32
Identities = 65/99 (65%), Positives = 74/99 (74%)
Frame = +1
Query: 38 TTSATDVYNQPFVIGVAGGAASGKKTVCDMIVQQLHDQRVVLVNQDSFYNNLTEEERARV 97
T + + NQPFVIGV+GG ASGK TVCDMI+QQLHD RVVLVNQDSFY L EE RV
Sbjct: 37 TLTLDSLPNQPFVIGVSGGTASGKTTVCDMIIQQLHDHRVVLVNQDSFYRGLNPEELERV 216
Query: 98 QDYNFDHPDAFDTEELLRVMDKLKHGEAVDIPKYDFKSY 136
+YNFDHPDAFDTE+LL KL G+ V +P YDFK +
Sbjct: 217 HEYNFDHPDAFDTEQLLECTRKLISGQGVHVPIYDFKKH 333
>AW164847
Length = 298
Score = 114 bits (285), Expect = 1e-25
Identities = 57/60 (95%), Positives = 58/60 (96%), Gaps = 1/60 (1%)
Frame = +3
Query: 418 ISAPKGVHVVCKCFPRIKIVTSEIEIGLNEDFRVIPGMGEFGDRYFGT-DDDEQVVVASQ 476
ISAPKGVHVVCK FPRIKIVTSEIEIGLNEDFRVIPGMGEFGDRYFGT DDDEQVVV+SQ
Sbjct: 3 ISAPKGVHVVCKSFPRIKIVTSEIEIGLNEDFRVIPGMGEFGDRYFGTDDDDEQVVVSSQ 182
>AW756633 homologue to GP|8778301|gb|A F14J16.5 {Arabidopsis thaliana},
partial (10%)
Length = 330
Score = 111 bits (277), Expect = 8e-25
Identities = 52/53 (98%), Positives = 53/53 (99%)
Frame = +3
Query: 316 GSVYTGVDFCKRLCGVSVIRSGESMENALRACCKGIKIGKILIHREGDNGQQL 368
GSVYTGVDFCKRLCGVSVIRSGESMENALRACCKGIKIGKILIHREGDNGQQ+
Sbjct: 102 GSVYTGVDFCKRLCGVSVIRSGESMENALRACCKGIKIGKILIHREGDNGQQV 260
>AW459115 weakly similar to GP|10177966|db uridine kinase-like protein
{Arabidopsis thaliana}, partial (25%)
Length = 426
Score = 102 bits (253), Expect = 5e-22
Identities = 47/99 (47%), Positives = 73/99 (73%), Gaps = 2/99 (2%)
Frame = +1
Query: 89 LTEEERARVQDYNFDHPDAFDTEELLRVMDKLKHGEAVDIPKYDFKSYK--SDALRRVNP 146
L EE RV YNF+HPDAFDTE+LL KL G++V +P Y+FK ++ S++ R +N
Sbjct: 13 LNPEELERVHKYNFNHPDAFDTEQLLECTMKLISGQSVHVPIYNFKKHQHSSNSFRPINA 192
Query: 147 SDVIILEGILVFHDPRVRELMNMKIFVDTDADVRLARRI 185
S++IIL+GIL+FH+ ++ L+N+KIF++TD +++L +I
Sbjct: 193 SNIIILKGILIFHNXHIQNLINIKIFINTDTNIKLTHKI 309
Score = 60.8 bits (146), Expect = 1e-09
Identities = 25/39 (64%), Positives = 33/39 (84%)
Frame = +3
Query: 231 DNHVAVDLIVQHIRTKLGQHDLCKIYPNLYVIHSTFQIR 269
+NH+ ++LI+QHI TKL QH+L KIYPN Y+I STFQI+
Sbjct: 309 NNHITINLIIQHIHTKLHQHNLYKIYPNTYIIQSTFQIK 425
>BG650864
Length = 381
Score = 97.1 bits (240), Expect = 2e-20
Identities = 46/55 (83%), Positives = 50/55 (90%)
Frame = +3
Query: 259 LYVIHSTFQIRGMHTLIRDAQITKHDFVFYSDRLIRLVVEHGLGHLPFTEKQVIT 313
+ I ++ QIRGMHTLIRD+Q TKHDFVFYSDRLIRLVVEHGLGHLPFTEKQVIT
Sbjct: 216 IITIIASMQIRGMHTLIRDSQTTKHDFVFYSDRLIRLVVEHGLGHLPFTEKQVIT 380
Score = 81.6 bits (200), Expect = 7e-16
Identities = 36/39 (92%), Positives = 38/39 (97%)
Frame = +2
Query: 230 GDNHVAVDLIVQHIRTKLGQHDLCKIYPNLYVIHSTFQI 268
GDNHVA+DLIVQHIRTKLGQHDLCKIYPNLYVI STFQ+
Sbjct: 2 GDNHVAIDLIVQHIRTKLGQHDLCKIYPNLYVIQSTFQV 118
>BE806441 similar to GP|9294212|dbj| uridine kinase-like protein {Arabidopsis
thaliana}, partial (13%)
Length = 395
Score = 83.2 bits (204), Expect = 2e-16
Identities = 44/80 (55%), Positives = 53/80 (66%), Gaps = 6/80 (7%)
Frame = +3
Query: 10 LMGSSSEVHFSGFHMDGFEKREASIEQPTTSATDVY------NQPFVIGVAGGAASGKKT 63
+M ++S HFSG +DG A+ ++SA NQPFVIGV+GG ASGK T
Sbjct: 156 VMEAASGPHFSGLRLDGRMPSSATAAAASSSADSTLTPDFLPNQPFVIGVSGGTASGKTT 335
Query: 64 VCDMIVQQLHDQRVVLVNQD 83
VCDMI+QQLHD RVVLVNQD
Sbjct: 336 VCDMIIQQLHDHRVVLVNQD 395
>TC205803 similar to UP|UPP_TOBAC (P93394) Uracil phosphoribosyltransferase
(UMP pyrophosphorylase) (UPRTase) , partial (96%)
Length = 1173
Score = 82.4 bits (202), Expect = 4e-16
Identities = 54/175 (30%), Positives = 91/175 (51%), Gaps = 3/175 (1%)
Frame = +1
Query: 295 LVVEHGLGHLPFTEKQVITPTGSVYTG-VDFCKRLCGVSVIRSGESMENALRACCKGIKI 353
L+ E LP ++ +P G +D + + + ++R+G ++ + IK
Sbjct: 424 LMYEASRDWLPTVSGEIQSPMGVASVELIDPREPVAVIPILRAGLALAEHASSILPAIKT 603
Query: 354 GKILIHREGDNGQQLIY-EKLPNDISE-RHVLLLDPILGTGNSAVQAISLLIQKGVPESN 411
+ I R+ + Q IY KLP E V ++DP+L TG + V A+SLL ++GV
Sbjct: 604 YHLGISRDEETLQPTIYLNKLPEKFPEGSKVFVVDPMLATGGTIVAAVSLLKERGVGNKQ 783
Query: 412 IIFLNLISAPKGVHVVCKCFPRIKIVTSEIEIGLNEDFRVIPGMGEFGDRYFGTD 466
I ++ +SAP + + + FP + + T I+ +NE +IPG+G+ GDR FGTD
Sbjct: 784 IKVISAVSAPPALQKLSEQFPGLHVYTGIIDPEVNEKGFIIPGLGDAGDRSFGTD 948
>BI424501
Length = 239
Score = 72.8 bits (177), Expect = 3e-13
Identities = 35/40 (87%), Positives = 38/40 (94%)
Frame = +3
Query: 4 AKSAVDLMGSSSEVHFSGFHMDGFEKREASIEQPTTSATD 43
AKSAV+LM SSSEVHFSGFHMDGFE+R+ASIEQPTTS TD
Sbjct: 120 AKSAVELMESSSEVHFSGFHMDGFEQRKASIEQPTTSETD 239
>TC225348 homologue to UP|KPPR_MESCR (P27774) Phosphoribulokinase,
chloroplast precursor (Phosphopentokinase) (PRKASE)
(PRK) , partial (91%)
Length = 1776
Score = 69.7 bits (169), Expect = 3e-12
Identities = 44/157 (28%), Positives = 83/157 (52%)
Frame = +3
Query: 105 PDAFDTEELLRVMDKLKHGEAVDIPKYDFKSYKSDALRRVNPSDVIILEGILVFHDPRVR 164
P A D + + + LK G AV+ P Y+ + D + P ++++EG+ +D RVR
Sbjct: 621 PRANDFDLMYEQVKALKDGIAVEKPIYNHVTGLLDPPELIKPPKILVIEGLHPMYDSRVR 800
Query: 165 ELMNMKIFVDTDADVRLARRIRRDTSEKDRDIGAILDQYSKFVKPAFDDFILPTKKYADI 224
+L++ I++D +V+ A +I+RD +E+ + +I + KP F+ +I P K+YAD
Sbjct: 801 DLLDFSIYLDISNEVKFAWKIQRDMAERGHSLESIKASI-EARKPDFEAYIDPQKQYADA 977
Query: 225 IIPRGGDNHVAVDLIVQHIRTKLGQHDLCKIYPNLYV 261
+I + D + +R +L Q + K + +Y+
Sbjct: 978 VIEVLPTQLIPDDNEGKILRVRLIQKEGVKYFSPVYL 1088
>TC225351 homologue to UP|P93681 (P93681) Phosphoribulokinase (Fragment) ,
partial (55%)
Length = 656
Score = 63.5 bits (153), Expect = 2e-10
Identities = 39/142 (27%), Positives = 75/142 (52%)
Frame = +2
Query: 120 LKHGEAVDIPKYDFKSYKSDALRRVNPSDVIILEGILVFHDPRVRELMNMKIFVDTDADV 179
LK V+ P Y+ + D + P ++++EG+ +D RVR+L++ I++D +V
Sbjct: 44 LKDXXXVEKPIYNHVTGLLDPPELIKPPKILVIEGLHPMYDSRVRDLLDFSIYLDISNEV 223
Query: 180 RLARRIRRDTSEKDRDIGAILDQYSKFVKPAFDDFILPTKKYADIIIPRGGDNHVAVDLI 239
+ A +I+RD +E+ + +I + KP F+ +I P K+YAD +I + D
Sbjct: 224 KFAWKIQRDMAERGHSLESIKASI-EARKPDFEAYIDPQKQYADAVIEVLPTQLIPDDNE 400
Query: 240 VQHIRTKLGQHDLCKIYPNLYV 261
+ +R +L Q + K + +Y+
Sbjct: 401 GKILRVRLIQKEGVKYFSPVYL 466
>AW459831
Length = 396
Score = 61.2 bits (147), Expect = 1e-09
Identities = 26/33 (78%), Positives = 32/33 (96%)
Frame = +2
Query: 435 KIVTSEIEIGLNEDFRVIPGMGEFGDRYFGTDD 467
+IVTSEI+I +NE++RVIPG+GEFGDRYFGTDD
Sbjct: 5 EIVTSEIDIEINEEYRVIPGLGEFGDRYFGTDD 103
>BM953841 similar to GP|7939517|dbj| uridine kinase-like protein {Arabidopsis
thaliana}, partial (6%)
Length = 421
Score = 55.1 bits (131), Expect = 7e-08
Identities = 24/28 (85%), Positives = 26/28 (92%)
Frame = +3
Query: 308 EKQVITPTGSVYTGVDFCKRLCGVSVIR 335
EKQV TPT SVY+GVDFCKRLCGVS+IR
Sbjct: 3 EKQVTTPTASVYSGVDFCKRLCGVSIIR 86
>AW350036 homologue to SP|P25697|KPPR Phosphoribulokinase chloroplast
precursor (EC 2.7.1.19) (Phosphopentokinase) (PRKASE)
(PRK)., partial (28%)
Length = 590
Score = 52.8 bits (125), Expect = 3e-07
Identities = 32/113 (28%), Positives = 59/113 (51%)
Frame = -2
Query: 107 AFDTEELLRVMDKLKHGEAVDIPKYDFKSYKSDALRRVNPSDVIILEGILVFHDPRVREL 166
A D + + + K G AV+ P Y+ + D + P ++EG+ +D RVR+L
Sbjct: 589 ANDFDLMYEQVKAXKDGIAVEKPIYNHVTGLLDPPELIKPPKXXVIEGLHPMYDSRVRDL 410
Query: 167 MNMKIFVDTDADVRLARRIRRDTSEKDRDIGAILDQYSKFVKPAFDDFILPTK 219
++ I++D +V+ A +I+RD +E+ + +I + KP F+ +I P K
Sbjct: 409 LDFSIYLDISNEVKFAWKIQRDMAERGHSLESIKASI-EARKPDFEAYIDPQK 254
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.141 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,268,711
Number of Sequences: 63676
Number of extensions: 241141
Number of successful extensions: 976
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 969
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 970
length of query: 476
length of database: 12,639,632
effective HSP length: 101
effective length of query: 375
effective length of database: 6,208,356
effective search space: 2328133500
effective search space used: 2328133500
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC147712.11


