

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147537.6 + phase: 0
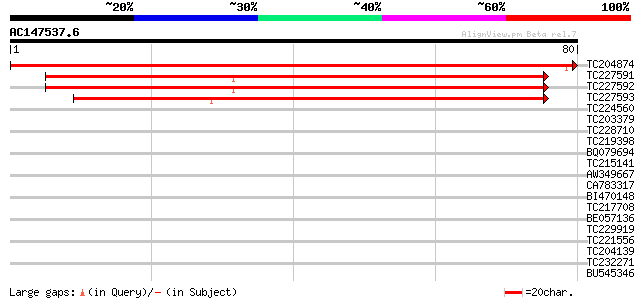
(80 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204874 similar to UP|Q9M206 (Q9M206) Transport protein subunit... 138 4e-34
TC227591 homologue to GB|BAC42746.1|26451290|AK118120 protein tr... 96 4e-21
TC227592 homologue to GB|BAC42746.1|26451290|AK118120 protein tr... 96 4e-21
TC227593 similar to GB|BAC42746.1|26451290|AK118120 protein tran... 92 6e-20
TC224560 similar to UP|O24292 (O24292) CP12 precursor, partial (... 28 1.1
TC203379 similar to GB|BAC55960.1|28071265|AB087408 actin bindin... 28 1.1
TC228710 similar to UP|Q6NQ98 (Q6NQ98) At5g58020, partial (41%) 27 1.4
TC219398 similar to UP|Q7U2T0 (Q7U2T0) PE-PGRS family protein, p... 27 1.9
BQ079694 27 1.9
TC215141 similar to GB|AAL32703.1|17065098|AY062625 nucleotide s... 27 2.4
AW349667 27 2.4
CA783317 similar to GP|6523547|emb| hydroxyproline-rich glycopro... 27 2.4
BI470148 similar to GP|2352260|gb|A keratin {Canis familiaris}, ... 27 2.4
TC217708 similar to UP|O65896 (O65896) Cytidine deaminase 1 (At... 27 2.4
BE057136 similar to PIR|JQ1060|JQ10 glycine-rich protein 1 - Ara... 26 3.2
TC229919 weakly similar to UP|O64855 (O64855) Expressed protein ... 26 3.2
TC221556 weakly similar to UP|Q9FGH9 (Q9FGH9) Leucine zipper pro... 26 3.2
TC204139 similar to UP|O04002 (O04002) CDSP32 protein (Chloropla... 26 4.2
TC232271 similar to UP|Q8RWR1 (Q8RWR1) AT3g20810/MOE17_10, parti... 26 4.2
BU545346 25 7.1
>TC204874 similar to UP|Q9M206 (Q9M206) Transport protein subunit-like,
partial (93%)
Length = 1239
Score = 138 bits (348), Expect = 4e-34
Identities = 72/82 (87%), Positives = 75/82 (90%), Gaps = 2/82 (2%)
Frame = +3
Query: 1 MALGGGAPQRGSAAATASMRRRKTAGGGASGGAAGTMLQFYTDDAPGLKISPNVVLVMSI 60
MALGG AP RGSAAATASMRRR+T GG ASGGAAGTMLQFYTDDAPGLKISPNVVLVMSI
Sbjct: 618 MALGGTAPPRGSAAATASMRRRRTTGGAASGGAAGTMLQFYTDDAPGLKISPNVVLVMSI 797
Query: 61 GFIAFVAVLHVVGKLYL--REA 80
GFIAFVA+LHV+GKLY REA
Sbjct: 798 GFIAFVAILHVMGKLYFVRREA 863
>TC227591 homologue to GB|BAC42746.1|26451290|AK118120 protein transport
protein subunit - like {Arabidopsis thaliana;} , partial
(80%)
Length = 541
Score = 95.5 bits (236), Expect = 4e-21
Identities = 50/76 (65%), Positives = 55/76 (71%), Gaps = 5/76 (6%)
Frame = +1
Query: 6 GAPQRGSAAATASMRRRKTAGGGAS-----GGAAGTMLQFYTDDAPGLKISPNVVLVMSI 60
G RGSAAATA MRRR+ GG +S GG + ML+FYTDDAPGLKISP VVLVMS+
Sbjct: 145 GMAPRGSAAATAGMRRRRLGGGNSSASTGVGGGSSNMLRFYTDDAPGLKISPTVVLVMSL 324
Query: 61 GFIAFVAVLHVVGKLY 76
FI FV LHV GKLY
Sbjct: 325 CFIGFVTALHVFGKLY 372
>TC227592 homologue to GB|BAC42746.1|26451290|AK118120 protein transport
protein subunit - like {Arabidopsis thaliana;} , partial
(80%)
Length = 574
Score = 95.5 bits (236), Expect = 4e-21
Identities = 50/76 (65%), Positives = 55/76 (71%), Gaps = 5/76 (6%)
Frame = +3
Query: 6 GAPQRGSAAATASMRRRKTAGGGAS-----GGAAGTMLQFYTDDAPGLKISPNVVLVMSI 60
G RGSAAATA MRRR+ GG +S GG + ML+FYTDDAPGLKISP VVLVMS+
Sbjct: 135 GMAPRGSAAATAGMRRRRLGGGNSSASTGVGGGSSNMLRFYTDDAPGLKISPTVVLVMSL 314
Query: 61 GFIAFVAVLHVVGKLY 76
FI FV LHV GKLY
Sbjct: 315 CFIGFVTALHVFGKLY 362
>TC227593 similar to GB|BAC42746.1|26451290|AK118120 protein transport
protein subunit - like {Arabidopsis thaliana;} , partial
(76%)
Length = 515
Score = 91.7 bits (226), Expect = 6e-20
Identities = 48/74 (64%), Positives = 52/74 (69%), Gaps = 7/74 (9%)
Frame = +1
Query: 10 RGSAAATASMRRRKTAGG-------GASGGAAGTMLQFYTDDAPGLKISPNVVLVMSIGF 62
RG+AAATA MRRR+ GG G SG ML+FYTDDAPGLKISP VVLVMS+ F
Sbjct: 145 RGTAAATAGMRRRRLGGGTSSASVSGGSGAGGSNMLRFYTDDAPGLKISPTVVLVMSLCF 324
Query: 63 IAFVAVLHVVGKLY 76
I FV LHV GKLY
Sbjct: 325 IGFVTALHVFGKLY 366
>TC224560 similar to UP|O24292 (O24292) CP12 precursor, partial (56%)
Length = 689
Score = 27.7 bits (60), Expect = 1.1
Identities = 13/31 (41%), Positives = 14/31 (44%)
Frame = +2
Query: 4 GGGAPQRGSAAATASMRRRKTAGGGASGGAA 34
GGG Q S RRR+ GG GG A
Sbjct: 365 GGGEHQERGGGVLGSWRRRRVRGGVGRGGGA 457
>TC203379 similar to GB|BAC55960.1|28071265|AB087408 actin binding protein
{Arabidopsis thaliana;} , partial (29%)
Length = 1246
Score = 27.7 bits (60), Expect = 1.1
Identities = 15/36 (41%), Positives = 16/36 (43%)
Frame = -2
Query: 4 GGGAPQRGSAAATASMRRRKTAGGGASGGAAGTMLQ 39
GGG P G A GGG GGA GT L+
Sbjct: 108 GGGGPPGGGGGGGAP------GGGGGGGGADGTSLE 19
>TC228710 similar to UP|Q6NQ98 (Q6NQ98) At5g58020, partial (41%)
Length = 846
Score = 27.3 bits (59), Expect = 1.4
Identities = 18/44 (40%), Positives = 21/44 (46%), Gaps = 5/44 (11%)
Frame = +3
Query: 15 ATASMRRRKTAGGGASGGAAGT-----MLQFYTDDAPGLKISPN 53
+T S+R R GGG GGA G L Y D P K+ PN
Sbjct: 297 STLSLRSR-LLGGGGDGGATGAESRDCYLNMYADKKPD-KVDPN 422
>TC219398 similar to UP|Q7U2T0 (Q7U2T0) PE-PGRS family protein, partial (4%)
Length = 612
Score = 26.9 bits (58), Expect = 1.9
Identities = 15/35 (42%), Positives = 16/35 (44%)
Frame = +1
Query: 2 ALGGGAPQRGSAAATASMRRRKTAGGGASGGAAGT 36
A GG A GSAA R GGGA G G+
Sbjct: 79 ASGGVAGTGGSAAVNGERERIGVRGGGAGGCGCGS 183
>BQ079694
Length = 426
Score = 26.9 bits (58), Expect = 1.9
Identities = 13/32 (40%), Positives = 17/32 (52%)
Frame = -3
Query: 4 GGGAPQRGSAAATASMRRRKTAGGGASGGAAG 35
GGG R SAA +R R+++ G S G G
Sbjct: 349 GGGGETRNSAATPTRVRGRRSSRPGRSRGGEG 254
>TC215141 similar to GB|AAL32703.1|17065098|AY062625 nucleotide sugar
epimerase-like protein {Arabidopsis thaliana;} , partial
(89%)
Length = 1743
Score = 26.6 bits (57), Expect = 2.4
Identities = 12/32 (37%), Positives = 15/32 (46%)
Frame = -3
Query: 4 GGGAPQRGSAAATASMRRRKTAGGGASGGAAG 35
GGG G A+ RRR+ + G GG G
Sbjct: 298 GGGRRSGGPASPRRRRRRRRGSSGSGRGGGGG 203
Score = 25.8 bits (55), Expect = 4.2
Identities = 11/32 (34%), Positives = 15/32 (46%)
Frame = -3
Query: 4 GGGAPQRGSAAATASMRRRKTAGGGASGGAAG 35
GGG + G A+ RRR+ G+ G G
Sbjct: 301 GGGGRRSGGPASPRRRRRRRRGSSGSGRGGGG 206
>AW349667
Length = 148
Score = 26.6 bits (57), Expect = 2.4
Identities = 11/14 (78%), Positives = 12/14 (85%)
Frame = -3
Query: 63 IAFVAVLHVVGKLY 76
IAFV LHV+GKLY
Sbjct: 146 IAFVTALHVLGKLY 105
>CA783317 similar to GP|6523547|emb| hydroxyproline-rich glycoprotein DZ-HRGP
{Volvox carteri f. nagariensis}, partial (23%)
Length = 159
Score = 26.6 bits (57), Expect = 2.4
Identities = 12/29 (41%), Positives = 12/29 (41%)
Frame = +1
Query: 4 GGGAPQRGSAAATASMRRRKTAGGGASGG 32
GGG G RRR GGG GG
Sbjct: 73 GGGGGGGGGGGGXXEXRRRGGGGGGKGGG 159
Score = 25.0 bits (53), Expect = 7.1
Identities = 11/29 (37%), Positives = 12/29 (40%)
Frame = +1
Query: 4 GGGAPQRGSAAATASMRRRKTAGGGASGG 32
GGG G RR+ GGG GG
Sbjct: 70 GGGGGGGGGGGGGXXEXRRRGGGGGGKGG 156
>BI470148 similar to GP|2352260|gb|A keratin {Canis familiaris}, partial (4%)
Length = 413
Score = 26.6 bits (57), Expect = 2.4
Identities = 16/37 (43%), Positives = 18/37 (48%), Gaps = 6/37 (16%)
Frame = -1
Query: 4 GGGAPQRG------SAAATASMRRRKTAGGGASGGAA 34
GGG+ RG AAA RRR + G G G AA
Sbjct: 356 GGGSGSRGVEYLRRDAAAGGENRRRSSGGRGGGGEAA 246
>TC217708 similar to UP|O65896 (O65896) Cytidine deaminase 1 (At2g19570) ,
partial (69%)
Length = 954
Score = 26.6 bits (57), Expect = 2.4
Identities = 12/27 (44%), Positives = 16/27 (58%)
Frame = -1
Query: 5 GGAPQRGSAAATASMRRRKTAGGGASG 31
GGAP G ++RR ++ GGGA G
Sbjct: 279 GGAPHSGRGW**GTVRRGRSGGGGAQG 199
>BE057136 similar to PIR|JQ1060|JQ10 glycine-rich protein 1 - Arabidopsis
thaliana (fragment), partial (19%)
Length = 420
Score = 26.2 bits (56), Expect = 3.2
Identities = 15/33 (45%), Positives = 16/33 (48%)
Frame = -3
Query: 3 LGGGAPQRGSAAATASMRRRKTAGGGASGGAAG 35
LGGGA A A + AGGGA GG G
Sbjct: 121 LGGGAGGGFGGGAGAGIGGGGGAGGGAGGGFGG 23
>TC229919 weakly similar to UP|O64855 (O64855) Expressed protein
(At2g44200/F6E13.34), partial (19%)
Length = 758
Score = 26.2 bits (56), Expect = 3.2
Identities = 13/27 (48%), Positives = 16/27 (59%)
Frame = +3
Query: 6 GAPQRGSAAATASMRRRKTAGGGASGG 32
GA + GSA+ S+RRR G SGG
Sbjct: 306 GAAEGGSASIAESVRRRTYYSQGRSGG 386
>TC221556 weakly similar to UP|Q9FGH9 (Q9FGH9) Leucine zipper protein
(AT5g58430/mqj2_20), partial (25%)
Length = 700
Score = 26.2 bits (56), Expect = 3.2
Identities = 12/23 (52%), Positives = 13/23 (56%)
Frame = +3
Query: 21 RRKTAGGGASGGAAGTMLQFYTD 43
RR+ GG SGGAAGT D
Sbjct: 555 RRRRRGGADSGGAAGTDFDIVID 623
>TC204139 similar to UP|O04002 (O04002) CDSP32 protein (Chloroplast
Drought-induced Stress Protein of 32kDa), partial (77%)
Length = 1243
Score = 25.8 bits (55), Expect = 4.2
Identities = 10/22 (45%), Positives = 14/22 (63%)
Frame = +1
Query: 7 APQRGSAAATASMRRRKTAGGG 28
+PQRG A +R+R+ GGG
Sbjct: 274 SPQRGGVRVGARIRQRQACGGG 339
>TC232271 similar to UP|Q8RWR1 (Q8RWR1) AT3g20810/MOE17_10, partial (24%)
Length = 729
Score = 25.8 bits (55), Expect = 4.2
Identities = 16/40 (40%), Positives = 17/40 (42%), Gaps = 8/40 (20%)
Frame = +1
Query: 4 GGGAPQRGSAA--------ATASMRRRKTAGGGASGGAAG 35
GGGA R A A RRR + GGG SG G
Sbjct: 199 GGGAAPRDLRAWRVRVREHGGAGFRRRHSRGGGGSGDGVG 318
>BU545346
Length = 539
Score = 25.0 bits (53), Expect = 7.1
Identities = 13/24 (54%), Positives = 17/24 (70%)
Frame = -1
Query: 5 GGAPQRGSAAATASMRRRKTAGGG 28
G PQ SA+AT SM ++T+GGG
Sbjct: 494 GSLPQLTSASAT-SMLMKRTSGGG 426
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.136 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,404,780
Number of Sequences: 63676
Number of extensions: 24375
Number of successful extensions: 503
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 402
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 486
length of query: 80
length of database: 12,639,632
effective HSP length: 56
effective length of query: 24
effective length of database: 9,073,776
effective search space: 217770624
effective search space used: 217770624
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 52 (24.6 bits)
Medicago: description of AC147537.6


