

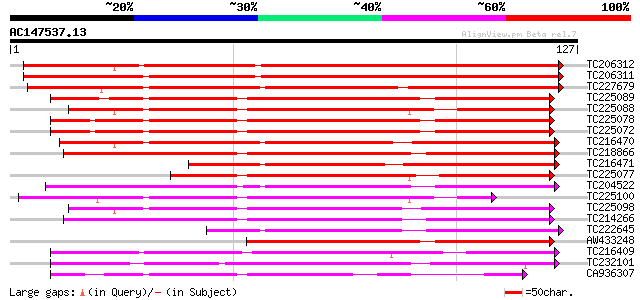
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147537.13 + phase: 0
(127 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC206312 weakly similar to UP|NLT1_PRUDU (Q43017) Nonspecific li... 155 5e-39
TC206311 similar to UP|Q8VX12 (Q8VX12) Lipid transfer protein pr... 150 1e-37
TC227679 similar to UP|NLT3_PRUDU (Q43019) Nonspecific lipid-tra... 135 7e-33
TC225089 similar to UP|Q93YX9 (Q93YX9) Lipid transfer protein, p... 104 1e-23
TC225088 similar to UP|Q6WAT8 (Q6WAT8) Lipid tranfer protein II,... 104 1e-23
TC225078 similar to UP|Q6WAT8 (Q6WAT8) Lipid tranfer protein II,... 104 1e-23
TC225072 similar to UP|Q6WAT8 (Q6WAT8) Lipid tranfer protein II,... 104 1e-23
TC216470 weakly similar to UP|Q9M6T9 (Q9M6T9) Lipid-transfer pro... 103 2e-23
TC218866 weakly similar to UP|NLTP_CICAR (O23758) Nonspecific li... 100 2e-22
TC216471 weakly similar to GB|CAB96874.1|8980444|MDO277164 mal d... 92 6e-20
TC225077 similar to UP|Q6WAT9 (Q6WAT9) Lipid transfer protein I,... 90 3e-19
TC204522 weakly similar to UP|Q93YX9 (Q93YX9) Lipid transfer pro... 89 7e-19
TC225100 weakly similar to UP|O24440 (O24440) Non-specific lipid... 86 3e-18
TC225098 weakly similar to UP|O24440 (O24440) Non-specific lipid... 84 2e-17
TC214266 weakly similar to UP|NLT1_ARATH (Q42589) Nonspecific li... 81 1e-16
TC222645 similar to PRF|2115353A.0|1098265|2115353A lipid transf... 69 6e-13
AW433248 similar to SP|Q9M5X6|NLTP_ Nonspecific lipid-transfer p... 69 8e-13
TC216409 similar to UP|Q9M7D7 (Q9M7D7) Lipid transfer protein, p... 68 1e-12
TC232101 weakly similar to UP|Q40454 (Q40454) Lipid transfer pro... 65 6e-12
CA936307 similar to PIR|T12079|T120 non-specific lipid transfer ... 65 1e-11
>TC206312 weakly similar to UP|NLT1_PRUDU (Q43017) Nonspecific lipid-transfer
protein 1 precursor (LTP 1), partial (51%)
Length = 767
Score = 155 bits (392), Expect = 5e-39
Identities = 76/125 (60%), Positives = 96/125 (76%), Gaps = 4/125 (3%)
Frame = +1
Query: 4 STMATRLVCLAIVCLVTFG----PKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCC 59
+++ TR+ +A++CL+ G P A+ V SCG VV++L PC+SY++ GG TVPA QCC
Sbjct: 100 ASLQTRVFLMALLCLLALGATIIPMAQGEV-SCGQVVSNLTPCISYVLYGGATVPA-QCC 273
Query: 60 NGIRNLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPN 119
NGIRNL MAQT DR+AVC CIKNAV SGF+Y++ NLNLAA LP+ CGVNIPYQISP+
Sbjct: 274 NGIRNLYGMAQTKPDRQAVCNCIKNAVRNSGFTYSHFNLNLAANLPKNCGVNIPYQISPD 453
Query: 120 TDCNR 124
TDC R
Sbjct: 454 TDCTR 468
>TC206311 similar to UP|Q8VX12 (Q8VX12) Lipid transfer protein precursor,
partial (26%)
Length = 915
Score = 150 bits (380), Expect = 1e-37
Identities = 71/121 (58%), Positives = 91/121 (74%)
Frame = +1
Query: 4 STMATRLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIR 63
++++TR+ + + + GP A+ +T CG VV++L PC+SY++ GG VP QCCNGIR
Sbjct: 271 ASLSTRIFFIMSLVCLALGPMAQGEMT-CGQVVSNLTPCISYVVYGGTNVPE-QCCNGIR 444
Query: 64 NLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCN 123
NL MAQT DR+AVC CIKN V SGF+Y++ NLNLAA LP+KCGVNIPYQISPNTDC
Sbjct: 445 NLYGMAQTKPDRQAVCNCIKNGVRNSGFNYSDFNLNLAANLPKKCGVNIPYQISPNTDCT 624
Query: 124 R 124
R
Sbjct: 625 R 627
>TC227679 similar to UP|NLT3_PRUDU (Q43019) Nonspecific lipid-transfer
protein 3 precursor (LTP 3), partial (40%)
Length = 659
Score = 135 bits (339), Expect = 7e-33
Identities = 69/124 (55%), Positives = 90/124 (71%), Gaps = 4/124 (3%)
Frame = +2
Query: 5 TMATRLVCLAIVCLV----TFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCN 60
++ +L+ +AIVCLV + PKA+AAVT C VV++L PC+SY++NGG TVP CCN
Sbjct: 86 SVVEKLLWMAIVCLVLGVISITPKAQAAVT-CNQVVSNLTPCISYVLNGGKTVPGP-CCN 259
Query: 61 GIRNLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNT 120
GI+ L +A +T DR+ VC CIKNAVS F Y N++ AA LP++CGVNIPYQISP+T
Sbjct: 260 GIKTLFNLAHSTPDRQTVCKCIKNAVS--AFHYGKSNVDRAAALPKQCGVNIPYQISPST 433
Query: 121 DCNR 124
DC R
Sbjct: 434 DCTR 445
>TC225089 similar to UP|Q93YX9 (Q93YX9) Lipid transfer protein, partial (78%)
Length = 793
Score = 104 bits (260), Expect = 1e-23
Identities = 53/113 (46%), Positives = 75/113 (65%)
Frame = +3
Query: 10 LVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTMA 69
++C+ + VT P A+AA+T CG V + PC+SY+ +GG P+ CCNG+++L+ A
Sbjct: 225 MMCMVVAMSVT--PMAQAAIT-CGQVAGDVSPCLSYLRSGGK--PSDACCNGVKSLSGAA 389
Query: 70 QTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDC 122
+TT DR+A C C+KN + G S LN AA LP KCGVNIPY+IS +T+C
Sbjct: 390 KTTADRQAACNCLKNLANNMGQS---LNAGNAASLPGKCGVNIPYKISTSTNC 539
>TC225088 similar to UP|Q6WAT8 (Q6WAT8) Lipid tranfer protein II, complete
Length = 1006
Score = 104 bits (259), Expect = 1e-23
Identities = 54/111 (48%), Positives = 74/111 (66%), Gaps = 2/111 (1%)
Frame = +2
Query: 14 AIVCLVTFG-PKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTMAQTT 72
A++C+V P A AA+T CG V +SL C+ Y+ GG P++ CCNG+++LN A+TT
Sbjct: 347 AVLCMVVVSAPMAHAAIT-CGQVTSSLANCIGYLQKGGT--PSSGCCNGVKSLNAAAKTT 517
Query: 73 NDRRAVCTCIKNAVSQ-SGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDC 122
DR+ C C+K+A SQ SGF N AA LP KCGV+IPY+IS +T+C
Sbjct: 518 ADRQTACNCLKSAASQISGFKANN-----AASLPGKCGVSIPYKISTSTNC 655
>TC225078 similar to UP|Q6WAT8 (Q6WAT8) Lipid tranfer protein II, partial
(90%)
Length = 697
Score = 104 bits (259), Expect = 1e-23
Identities = 52/113 (46%), Positives = 76/113 (67%)
Frame = +1
Query: 10 LVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTMA 69
++C+ +V + + P A+AA+T CG V + PC+SY+ +GG P+ CCNG+++L+ A
Sbjct: 121 VMCMVVVAM-SAAPMAQAAIT-CGQVAGDVSPCLSYLRSGGK--PSDACCNGVKSLSGAA 288
Query: 70 QTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDC 122
+TT DR+A C C+KN + G S LN AA LP KCGVNIPY+IS +T+C
Sbjct: 289 KTTADRQAACNCLKNLANNMGQS---LNAGNAASLPGKCGVNIPYKISTSTNC 438
>TC225072 similar to UP|Q6WAT8 (Q6WAT8) Lipid tranfer protein II, partial
(90%)
Length = 715
Score = 104 bits (259), Expect = 1e-23
Identities = 51/113 (45%), Positives = 76/113 (67%)
Frame = +1
Query: 10 LVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTMA 69
++C+ +V + + P A+AA+T CG V + PC SY+ +GG P+ CCNG+++L++ A
Sbjct: 124 VMCMVVVAM-SAAPMAQAAIT-CGQVAGDMSPCFSYLRSGGK--PSQACCNGVKSLSSAA 291
Query: 70 QTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDC 122
+TT DR+ C+C+KN + G S LN AA LP KCGVNIPY+IS +T+C
Sbjct: 292 KTTADRQGACSCLKNLANNMGQS---LNAGNAASLPGKCGVNIPYKISTSTNC 441
>TC216470 weakly similar to UP|Q9M6T9 (Q9M6T9) Lipid-transfer protein,
partial (61%)
Length = 648
Score = 103 bits (257), Expect = 2e-23
Identities = 50/115 (43%), Positives = 72/115 (62%), Gaps = 3/115 (2%)
Frame = +3
Query: 12 CLAIVCLVTFG---PKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTM 68
C+ +V + +FG P AEA + CG V ++ PC+ Y+ G VPA CCNG++++N
Sbjct: 111 CMVVVWMFSFGHIIPLAEAEIP-CGRVQITVAPCIGYLRGPGGGVPAP-CCNGVKSINNQ 284
Query: 69 AQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCN 123
A+TT DR+ VC C+K+ V S LNL + LP KCG+N+PY+I+P DCN
Sbjct: 285 AKTTPDRQGVCRCLKSTV----LSLAGLNLATLSALPSKCGINLPYKITPTIDCN 437
>TC218866 weakly similar to UP|NLTP_CICAR (O23758) Nonspecific lipid-transfer
protein precursor (LTP), partial (83%)
Length = 814
Score = 100 bits (248), Expect = 2e-22
Identities = 49/111 (44%), Positives = 68/111 (61%)
Frame = +1
Query: 13 LAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTMAQTT 72
+A++ +V A +CG V ++ PC+ Y+ GG P A+CCNG+RNL+ +A+TT
Sbjct: 103 VAVLYMVVVSAHMAHARINCGRVAVAVSPCLGYLRRGGR--PQARCCNGVRNLHALAKTT 276
Query: 73 NDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCN 123
DRR C C+K G +N N AA LPRKC VNIPY+IS +T+CN
Sbjct: 277 VDRRTACNCLKTFARGLG---RGVNANNAAALPRKCRVNIPYKISISTNCN 420
>TC216471 weakly similar to GB|CAB96874.1|8980444|MDO277164 mal d 3 {Malus x
domestica;} , partial (89%)
Length = 479
Score = 92.0 bits (227), Expect = 6e-20
Identities = 43/83 (51%), Positives = 54/83 (64%)
Frame = +3
Query: 41 PCVSYIMNGGNTVPAAQCCNGIRNLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNL 100
PC+ Y+ G VPAA CCNG+R++N A+TT DR+ VC C+K + S LNL
Sbjct: 18 PCIGYLRGPGGGVPAA-CCNGVRSINKEAKTTPDRQGVCRCLKT----TALSLPGLNLAT 182
Query: 101 AAGLPRKCGVNIPYQISPNTDCN 123
A LP KCGVN+PY+ISP DCN
Sbjct: 183 LAALPSKCGVNLPYKISPTIDCN 251
>TC225077 similar to UP|Q6WAT9 (Q6WAT9) Lipid transfer protein I, partial
(73%)
Length = 681
Score = 89.7 bits (221), Expect = 3e-19
Identities = 44/87 (50%), Positives = 60/87 (68%), Gaps = 1/87 (1%)
Frame = +3
Query: 37 TSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTMAQTTNDRRAVCTCIKNAVSQ-SGFSYTN 95
+SL C+ Y+ GG P++ CCNG+++LN A+TT DR+ C C+K+A SQ SGF
Sbjct: 210 SSLANCIGYLQKGGT--PSSGCCNGVKSLNAAAKTTADRQTACNCLKSAASQISGF---- 371
Query: 96 LNLNLAAGLPRKCGVNIPYQISPNTDC 122
N N AA LP KCGV+IPY+IS +T+C
Sbjct: 372 -NANNAASLPGKCGVSIPYKISTSTNC 449
>TC204522 weakly similar to UP|Q93YX9 (Q93YX9) Lipid transfer protein,
partial (27%)
Length = 881
Score = 88.6 bits (218), Expect = 7e-19
Identities = 41/115 (35%), Positives = 68/115 (58%)
Frame = +3
Query: 9 RLVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTM 68
++ C+ ++C+ SC V +L PC+SY+M GG+ VP + CC+G+RN+
Sbjct: 198 KVACVVLMCMAVMSAPMMVQAVSCNDVSVNLAPCLSYLMQGGD-VPES-CCSGVRNILGS 371
Query: 69 AQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCN 123
A TT D++ VC C++ A + G +N A LP +C V++PY+IS +T+C+
Sbjct: 372 ASTTFDKQTVCKCLQQAANNYG-----INDEYAQALPARCNVSVPYKISRSTNCD 521
>TC225100 weakly similar to UP|O24440 (O24440) Non-specific lipid transfer
protein PvLTP-24, partial (84%)
Length = 603
Score = 86.3 bits (212), Expect = 3e-18
Identities = 48/109 (44%), Positives = 62/109 (56%), Gaps = 2/109 (1%)
Frame = +1
Query: 3 YSTMATRLVCLAIVCL-VTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNG 61
Y T A++C+ P A AA+T CG V SL C+ Y+ NGG P + CCNG
Sbjct: 301 YMASLTGASLAAVLCMDAESAPMAHAAIT-CGQVTNSLINCIGYLQNGGT--PPSGCCNG 471
Query: 62 IRNLNTMAQTTNDRRAVCTCIKNAVSQ-SGFSYTNLNLNLAAGLPRKCG 109
+++LN A+TT DR+ C C+K+A SQ SGF N AA LP KCG
Sbjct: 472 VKSLNAAAKTTADRQTACNCLKSAASQISGFKANN-----AASLPGKCG 603
>TC225098 weakly similar to UP|O24440 (O24440) Non-specific lipid transfer
protein PvLTP-24, partial (97%)
Length = 717
Score = 84.0 bits (206), Expect = 2e-17
Identities = 45/110 (40%), Positives = 64/110 (57%), Gaps = 1/110 (0%)
Frame = +1
Query: 14 AIVCLVTFG-PKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTMAQTT 72
A++C+V P A AA+T CG V +SL C+ Y+ GG P++ CCNG+++LN + T
Sbjct: 400 AVLCMVVVSAPMAHAAIT-CGQVTSSLTNCIGYLQKGGT--PSSGCCNGVKSLNAATKIT 570
Query: 73 NDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDC 122
DR+ C C+K SQ + N A LP CGV I Y+IS +T+C
Sbjct: 571 ADRQTECNCLKLVASQ----IYSFKANNAVSLPG*CGVYILYKISTSTNC 708
>TC214266 weakly similar to UP|NLT1_ARATH (Q42589) Nonspecific lipid-transfer
protein 1 precursor (LTP 1), partial (36%)
Length = 699
Score = 81.3 bits (199), Expect = 1e-16
Identities = 39/110 (35%), Positives = 62/110 (55%)
Frame = +3
Query: 13 LAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTMAQTT 72
+ I+C+ G A +C V ++ C+SY+M GG P+ CC+G++N+ A TT
Sbjct: 108 VVIMCMAVVGAPMMAQAMTCNDVTVNMAQCLSYLMQGGT--PSTLCCSGVKNILGSAVTT 281
Query: 73 NDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDC 122
D++ VC C+K ++ N+N A LP C VN+PY+IS +T+C
Sbjct: 282 VDKQTVCNCLKADAAR-----YNINDQYAQALPGFCKVNVPYKISRSTNC 416
>TC222645 similar to PRF|2115353A.0|1098265|2115353A lipid transfer protein.
{Hordeum vulgare;} , partial (27%)
Length = 487
Score = 68.9 bits (167), Expect = 6e-13
Identities = 32/80 (40%), Positives = 47/80 (58%)
Frame = +2
Query: 45 YIMNGGNTVPAAQCCNGIRNLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGL 104
Y+++G PAA CC+G + L + A T+ D++A C CIK+ N+N LA L
Sbjct: 8 YLVSGSGQPPAA-CCSGAKALASAATTSEDKKAACNCIKSTSKS-----ININSQLAQAL 169
Query: 105 PRKCGVNIPYQISPNTDCNR 124
P CG+ +P ISPN DC++
Sbjct: 170 PGNCGITLPVAISPNADCSK 229
>AW433248 similar to SP|Q9M5X6|NLTP_ Nonspecific lipid-transfer protein
precursor (LTP) (Allergen Pyr c 3). [Pear] {Pyrus
communis}, partial (23%)
Length = 358
Score = 68.6 bits (166), Expect = 8e-13
Identities = 33/69 (47%), Positives = 44/69 (62%)
Frame = +2
Query: 54 PAAQCCNGIRNLNTMAQTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIP 113
P+ CCNG+++L++ A TT DR+ TC+ N + G S LN A LP KCGVNIP
Sbjct: 152 PSHACCNGLKSLSSAANTTYDRQRAYTCLINLANNMGQS---LNAGHAPSLPDKCGVNIP 322
Query: 114 YQISPNTDC 122
Y IS +T+C
Sbjct: 323 YMISTSTNC 349
>TC216409 similar to UP|Q9M7D7 (Q9M7D7) Lipid transfer protein, partial (87%)
Length = 762
Score = 67.8 bits (164), Expect = 1e-12
Identities = 37/115 (32%), Positives = 61/115 (52%), Gaps = 1/115 (0%)
Frame = +1
Query: 10 LVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTMA 69
LV + C++ AE+ + SC + L PC+ Y + GGN ++ CC G+ +LN
Sbjct: 97 LVASVMACMLITCSYAESTL-SCDQITIWLTPCIPYGVLGGNV--SSLCCQGVYSLNAAY 267
Query: 70 QTTNDRRAVCTCIKN-AVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQISPNTDCN 123
++ DRR C CIK+ A G YT +N + +CG P+++ P+T+C+
Sbjct: 268 KSAEDRRGACQCIKDRAACIPGIDYTRVN-----QVGPRCGSKCPFKVYPSTNCS 417
>TC232101 weakly similar to UP|Q40454 (Q40454) Lipid transfer protein,
partial (42%)
Length = 518
Score = 65.5 bits (158), Expect = 6e-12
Identities = 36/115 (31%), Positives = 60/115 (51%), Gaps = 1/115 (0%)
Frame = +2
Query: 10 LVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTMA 69
++ +AI+ + G A SC V+ + PC S++ GG+ P+ CCNGI+ L+ A
Sbjct: 23 IISIAILLITRSGALASI---SCATVIEEVAPCTSFLQ-GGSKQPSVACCNGIKKLSGEA 190
Query: 70 QTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPY-QISPNTDCN 123
T +R A+C C+K ++ G N + +P+ CG++ I NTDC+
Sbjct: 191 GTHQNRTAICQCLKEGLATIG----NYDPKRVPQVPKDCGLSATLPPIDKNTDCS 343
>CA936307 similar to PIR|T12079|T120 non-specific lipid transfer protein
LTP-24 drought and ABA induced - kidney bean, partial
(59%)
Length = 422
Score = 64.7 bits (156), Expect = 1e-11
Identities = 40/107 (37%), Positives = 56/107 (51%)
Frame = +3
Query: 10 LVCLAIVCLVTFGPKAEAAVTSCGPVVTSLYPCVSYIMNGGNTVPAAQCCNGIRNLNTMA 69
L+C+ +V P A AA+T CG V +SL PC+ YI GG+ P A CC+ +++LN A
Sbjct: 78 LLCMVVVS----APMAHAAIT-CGHVTSSLAPCIGYIQKGGS--PPAPCCS*VKSLNAAA 236
Query: 70 QTTNDRRAVCTCIKNAVSQSGFSYTNLNLNLAAGLPRKCGVNIPYQI 116
+TT DR+ + S+ G KCGV+IPY I
Sbjct: 237 KTTTDRQQ-----RRLASRQG----------------KCGVSIPYMI 314
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.134 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,759,943
Number of Sequences: 63676
Number of extensions: 90030
Number of successful extensions: 539
Number of sequences better than 10.0: 108
Number of HSP's better than 10.0 without gapping: 488
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 489
length of query: 127
length of database: 12,639,632
effective HSP length: 86
effective length of query: 41
effective length of database: 7,163,496
effective search space: 293703336
effective search space used: 293703336
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 53 (25.0 bits)
Medicago: description of AC147537.13


