

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147537.12 + phase: 0
(125 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
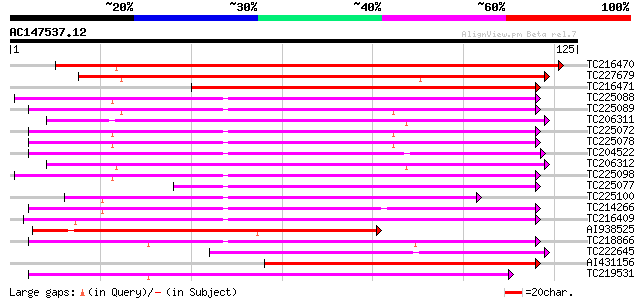
Score E
Sequences producing significant alignments: (bits) Value
TC216470 weakly similar to UP|Q9M6T9 (Q9M6T9) Lipid-transfer pro... 108 6e-25
TC227679 similar to UP|NLT3_PRUDU (Q43019) Nonspecific lipid-tra... 103 2e-23
TC216471 weakly similar to GB|CAB96874.1|8980444|MDO277164 mal d... 98 1e-21
TC225088 similar to UP|Q6WAT8 (Q6WAT8) Lipid tranfer protein II,... 97 2e-21
TC225089 similar to UP|Q93YX9 (Q93YX9) Lipid transfer protein, p... 95 9e-21
TC206311 similar to UP|Q8VX12 (Q8VX12) Lipid transfer protein pr... 93 4e-20
TC225072 similar to UP|Q6WAT8 (Q6WAT8) Lipid tranfer protein II,... 92 5e-20
TC225078 similar to UP|Q6WAT8 (Q6WAT8) Lipid tranfer protein II,... 92 5e-20
TC204522 weakly similar to UP|Q93YX9 (Q93YX9) Lipid transfer pro... 91 1e-19
TC206312 weakly similar to UP|NLT1_PRUDU (Q43017) Nonspecific li... 90 2e-19
TC225098 weakly similar to UP|O24440 (O24440) Non-specific lipid... 80 2e-16
TC225077 similar to UP|Q6WAT9 (Q6WAT9) Lipid transfer protein I,... 80 3e-16
TC225100 weakly similar to UP|O24440 (O24440) Non-specific lipid... 79 5e-16
TC214266 weakly similar to UP|NLT1_ARATH (Q42589) Nonspecific li... 79 5e-16
TC216409 similar to UP|Q9M7D7 (Q9M7D7) Lipid transfer protein, p... 75 1e-14
AI938525 similar to SP|Q43304|NLTD_ Nonspecific lipid-transfer p... 71 1e-13
TC218866 weakly similar to UP|NLTP_CICAR (O23758) Nonspecific li... 69 5e-13
TC222645 similar to PRF|2115353A.0|1098265|2115353A lipid transf... 68 9e-13
AI431156 similar to PIR|T12079|T120 non-specific lipid transfer ... 67 2e-12
TC219531 67 2e-12
>TC216470 weakly similar to UP|Q9M6T9 (Q9M6T9) Lipid-transfer protein,
partial (61%)
Length = 648
Score = 108 bits (270), Expect = 6e-25
Identities = 48/114 (42%), Positives = 71/114 (62%), Gaps = 2/114 (1%)
Frame = +3
Query: 11 TCLTMMMCMVLG--LPQTLDALSCLQVETKLMPCVPYVTGNGGYVPQPCCDGVKAINNQA 68
+C+ ++ G +P + C +V+ + PC+ Y+ G GG VP PCC+GVK+INNQA
Sbjct: 108 SCMVVVWMFSFGHIIPLAEAEIPCGRVQITVAPCIGYLRGPGGGVPAPCCNGVKSINNQA 287
Query: 69 VTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTCEKYVY 122
T DRQ CRC+K+ ++ GLN+ L+ LPSKCG++LPY + P+ C Y
Sbjct: 288 KTTPDRQGVCRCLKSTVLSLAGLNLATLSALPSKCGINLPYKITPTIDCNTVKY 449
>TC227679 similar to UP|NLT3_PRUDU (Q43019) Nonspecific lipid-transfer
protein 3 precursor (LTP 3), partial (40%)
Length = 659
Score = 103 bits (257), Expect = 2e-23
Identities = 49/110 (44%), Positives = 69/110 (62%), Gaps = 6/110 (5%)
Frame = +2
Query: 16 MMCMVLGL----PQTLDALSCLQVETKLMPCVPYVTGNGGYVPQPCCDGVKAINNQAVTK 71
++C+VLG+ P+ A++C QV + L PC+ YV G VP PCC+G+K + N A +
Sbjct: 116 IVCLVLGVISITPKAQAAVTCNQVVSNLTPCISYVLNGGKTVPGPCCNGIKTLFNLAHST 295
Query: 72 SDRQAACRCIKTATSAIH--GLNMDILAGLPSKCGVHLPYTLGPSTTCEK 119
DRQ C+CIK A SA H N+D A LP +CGV++PY + PST C +
Sbjct: 296 PDRQTVCKCIKNAVSAFHYGKSNVDRAAALPKQCGVNIPYQISPSTDCTR 445
>TC216471 weakly similar to GB|CAB96874.1|8980444|MDO277164 mal d 3 {Malus x
domestica;} , partial (89%)
Length = 479
Score = 97.8 bits (242), Expect = 1e-21
Identities = 41/77 (53%), Positives = 54/77 (69%)
Frame = +3
Query: 41 PCVPYVTGNGGYVPQPCCDGVKAINNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLP 100
PC+ Y+ G GG VP CC+GV++IN +A T DRQ CRC+KT ++ GLN+ LA LP
Sbjct: 18 PCIGYLRGPGGGVPAACCNGVRSINKEAKTTPDRQGVCRCLKTTALSLPGLNLATLAALP 197
Query: 101 SKCGVHLPYTLGPSTTC 117
SKCGV+LPY + P+ C
Sbjct: 198 SKCGVNLPYKISPTIDC 248
>TC225088 similar to UP|Q6WAT8 (Q6WAT8) Lipid tranfer protein II, complete
Length = 1006
Score = 97.1 bits (240), Expect = 2e-21
Identities = 50/117 (42%), Positives = 68/117 (57%), Gaps = 1/117 (0%)
Frame = +2
Query: 2 SSSMVLVKVTCLTMMMCMVL-GLPQTLDALSCLQVETKLMPCVPYVTGNGGYVPQPCCDG 60
S SM +KV L ++CMV+ P A++C QV + L C+ Y+ GG CC+G
Sbjct: 308 SVSMASLKVAFLAAVLCMVVVSAPMAHAAITCGQVTSSLANCIGYLQ-KGGTPSSGCCNG 484
Query: 61 VKAINNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTC 117
VK++N A T +DRQ AC C+K+A S I G + A LP KCGV +PY + ST C
Sbjct: 485 VKSLNAAAKTTADRQTACNCLKSAASQISGFKANNAASLPGKCGVSIPYKISTSTNC 655
>TC225089 similar to UP|Q93YX9 (Q93YX9) Lipid transfer protein, partial (78%)
Length = 793
Score = 94.7 bits (234), Expect = 9e-21
Identities = 49/117 (41%), Positives = 68/117 (57%), Gaps = 4/117 (3%)
Frame = +3
Query: 5 MVLVKVTCLTMMMCMVLGL---PQTLDALSCLQVETKLMPCVPYVTGNGGYVPQPCCDGV 61
M KV C+ MMCMV+ + P A++C QV + PC+ Y+ +GG CC+GV
Sbjct: 192 MTSFKVACVVAMMCMVVAMSVTPMAQAAITCGQVAGDVSPCLSYLR-SGGKPSDACCNGV 368
Query: 62 KAINNQAVTKSDRQAACRCIKT-ATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTC 117
K+++ A T +DRQAAC C+K A + LN A LP KCGV++PY + ST C
Sbjct: 369 KSLSGAAKTTADRQAACNCLKNLANNMGQSLNAGNAASLPGKCGVNIPYKISTSTNC 539
>TC206311 similar to UP|Q8VX12 (Q8VX12) Lipid transfer protein precursor,
partial (26%)
Length = 915
Score = 92.8 bits (229), Expect = 4e-20
Identities = 42/115 (36%), Positives = 65/115 (56%), Gaps = 4/115 (3%)
Frame = +1
Query: 9 KVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNGGYVPQPCCDGVKAINNQA 68
++ + ++C+ LG P ++C QV + L PC+ YV G VP+ CC+G++ + A
Sbjct: 286 RIFFIMSLVCLALG-PMAQGEMTCGQVVSNLTPCISYVVYGGTNVPEQCCNGIRNLYGMA 462
Query: 69 VTKSDRQAACRCIKTATS----AIHGLNMDILAGLPSKCGVHLPYTLGPSTTCEK 119
TK DRQA C CIK N+++ A LP KCGV++PY + P+T C +
Sbjct: 463 QTKPDRQAVCNCIKNGVRNSGFNYSDFNLNLAANLPKKCGVNIPYQISPNTDCTR 627
>TC225072 similar to UP|Q6WAT8 (Q6WAT8) Lipid tranfer protein II, partial
(90%)
Length = 715
Score = 92.4 bits (228), Expect = 5e-20
Identities = 48/118 (40%), Positives = 68/118 (56%), Gaps = 5/118 (4%)
Frame = +1
Query: 5 MVLVKVTCLTMMMCMVL----GLPQTLDALSCLQVETKLMPCVPYVTGNGGYVPQPCCDG 60
M +KV C+ +MCMV+ P A++C QV + PC Y+ +GG Q CC+G
Sbjct: 91 MASLKVACVVAVMCMVVVAMSAAPMAQAAITCGQVAGDMSPCFSYLR-SGGKPSQACCNG 267
Query: 61 VKAINNQAVTKSDRQAACRCIKT-ATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTC 117
VK++++ A T +DRQ AC C+K A + LN A LP KCGV++PY + ST C
Sbjct: 268 VKSLSSAAKTTADRQGACSCLKNLANNMGQSLNAGNAASLPGKCGVNIPYKISTSTNC 441
>TC225078 similar to UP|Q6WAT8 (Q6WAT8) Lipid tranfer protein II, partial
(90%)
Length = 697
Score = 92.4 bits (228), Expect = 5e-20
Identities = 48/118 (40%), Positives = 68/118 (56%), Gaps = 5/118 (4%)
Frame = +1
Query: 5 MVLVKVTCLTMMMCMVL----GLPQTLDALSCLQVETKLMPCVPYVTGNGGYVPQPCCDG 60
M +KV C+ +MCMV+ P A++C QV + PC+ Y+ +GG CC+G
Sbjct: 88 MASLKVACVVAVMCMVVVAMSAAPMAQAAITCGQVAGDVSPCLSYLR-SGGKPSDACCNG 264
Query: 61 VKAINNQAVTKSDRQAACRCIKT-ATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTC 117
VK+++ A T +DRQAAC C+K A + LN A LP KCGV++PY + ST C
Sbjct: 265 VKSLSGAAKTTADRQAACNCLKNLANNMGQSLNAGNAASLPGKCGVNIPYKISTSTNC 438
>TC204522 weakly similar to UP|Q93YX9 (Q93YX9) Lipid transfer protein,
partial (27%)
Length = 881
Score = 90.9 bits (224), Expect = 1e-19
Identities = 42/114 (36%), Positives = 65/114 (56%)
Frame = +3
Query: 5 MVLVKVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNGGYVPQPCCDGVKAI 64
M +KV C+ +M V+ P + A+SC V L PC+ Y+ GG VP+ CC GV+ I
Sbjct: 186 MASLKVACVVLMCMAVMSAPMMVQAVSCNDVSVNLAPCLSYLM-QGGDVPESCCSGVRNI 362
Query: 65 NNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTCE 118
A T D+Q C+C++ A + +G+N + LP++C V +PY + ST C+
Sbjct: 363 LGSASTTFDKQTVCKCLQQAANN-YGINDEYAQALPARCNVSVPYKISRSTNCD 521
>TC206312 weakly similar to UP|NLT1_PRUDU (Q43017) Nonspecific lipid-transfer
protein 1 precursor (LTP 1), partial (51%)
Length = 767
Score = 90.1 bits (222), Expect = 2e-19
Identities = 43/118 (36%), Positives = 63/118 (52%), Gaps = 7/118 (5%)
Frame = +1
Query: 9 KVTCLTMMMCMVLG---LPQTLDALSCLQVETKLMPCVPYVTGNGGYVPQPCCDGVKAIN 65
+V + ++ + LG +P +SC QV + L PC+ YV G VP CC+G++ +
Sbjct: 115 RVFLMALLCLLALGATIIPMAQGEVSCGQVVSNLTPCISYVLYGGATVPAQCCNGIRNLY 294
Query: 66 NQAVTKSDRQAACRCIKTATS----AIHGLNMDILAGLPSKCGVHLPYTLGPSTTCEK 119
A TK DRQA C CIK A N+++ A LP CGV++PY + P T C +
Sbjct: 295 GMAQTKPDRQAVCNCIKNAVRNSGFTYSHFNLNLAANLPKNCGVNIPYQISPDTDCTR 468
>TC225098 weakly similar to UP|O24440 (O24440) Non-specific lipid transfer
protein PvLTP-24, partial (97%)
Length = 717
Score = 80.1 bits (196), Expect = 2e-16
Identities = 42/117 (35%), Positives = 61/117 (51%), Gaps = 1/117 (0%)
Frame = +1
Query: 2 SSSMVLVKVTCLTMMMCMVL-GLPQTLDALSCLQVETKLMPCVPYVTGNGGYVPQPCCDG 60
S SM +KV L ++CMV+ P A++C QV + L C+ Y+ GG CC+G
Sbjct: 361 SVSMASLKVAFLAAVLCMVVVSAPMAHAAITCGQVTSSLTNCIGYLQ-KGGTPSSGCCNG 537
Query: 61 VKAINNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTC 117
VK++N +DRQ C C+K S I+ + LP CGV++ Y + ST C
Sbjct: 538 VKSLNAATKITADRQTECNCLKLVASQIYSFKANNAVSLPG*CGVYILYKISTSTNC 708
>TC225077 similar to UP|Q6WAT9 (Q6WAT9) Lipid transfer protein I, partial
(73%)
Length = 681
Score = 79.7 bits (195), Expect = 3e-16
Identities = 37/81 (45%), Positives = 49/81 (59%)
Frame = +3
Query: 37 TKLMPCVPYVTGNGGYVPQPCCDGVKAINNQAVTKSDRQAACRCIKTATSAIHGLNMDIL 96
+ L C+ Y+ GG CC+GVK++N A T +DRQ AC C+K+A S I G N +
Sbjct: 210 SSLANCIGYLQ-KGGTPSSGCCNGVKSLNAAAKTTADRQTACNCLKSAASQISGFNANNA 386
Query: 97 AGLPSKCGVHLPYTLGPSTTC 117
A LP KCGV +PY + ST C
Sbjct: 387 ASLPGKCGVSIPYKISTSTNC 449
>TC225100 weakly similar to UP|O24440 (O24440) Non-specific lipid transfer
protein PvLTP-24, partial (84%)
Length = 603
Score = 79.0 bits (193), Expect = 5e-16
Identities = 40/93 (43%), Positives = 54/93 (58%), Gaps = 1/93 (1%)
Frame = +1
Query: 13 LTMMMCM-VLGLPQTLDALSCLQVETKLMPCVPYVTGNGGYVPQPCCDGVKAINNQAVTK 71
L ++CM P A++C QV L+ C+ Y+ NGG P CC+GVK++N A T
Sbjct: 328 LAAVLCMDAESAPMAHAAITCGQVTNSLINCIGYLQ-NGGTPPSGCCNGVKSLNAAAKTT 504
Query: 72 SDRQAACRCIKTATSAIHGLNMDILAGLPSKCG 104
+DRQ AC C+K+A S I G + A LP KCG
Sbjct: 505 ADRQTACNCLKSAASQISGFKANNAASLPGKCG 603
>TC214266 weakly similar to UP|NLT1_ARATH (Q42589) Nonspecific lipid-transfer
protein 1 precursor (LTP 1), partial (36%)
Length = 699
Score = 79.0 bits (193), Expect = 5e-16
Identities = 43/114 (37%), Positives = 61/114 (52%), Gaps = 1/114 (0%)
Frame = +3
Query: 5 MVLVKVTCLTMMMCM-VLGLPQTLDALSCLQVETKLMPCVPYVTGNGGYVPQPCCDGVKA 63
M +KVT + ++MCM V+G P A++C V + C+ Y+ GG CC GVK
Sbjct: 81 MASMKVTWVVVIMCMAVVGAPMMAQAMTCNDVTVNMAQCLSYLM-QGGTPSTLCCSGVKN 257
Query: 64 INNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTC 117
I AVT D+Q C C+K A +A + +N LP C V++PY + ST C
Sbjct: 258 ILGSAVTTVDKQTVCNCLK-ADAARYNINDQYAQALPGFCKVNVPYKISRSTNC 416
>TC216409 similar to UP|Q9M7D7 (Q9M7D7) Lipid transfer protein, partial (87%)
Length = 762
Score = 74.7 bits (182), Expect = 1e-14
Identities = 40/115 (34%), Positives = 62/115 (53%), Gaps = 1/115 (0%)
Frame = +1
Query: 4 SMVLVKVTCL-TMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNGGYVPQPCCDGVK 62
+M + KVT + ++M CM++ LSC Q+ L PC+PY GG V CC GV
Sbjct: 73 TMAVNKVTLVASVMACMLITCSYAESTLSCDQITIWLTPCIPYGV-LGGNVSSLCCQGVY 249
Query: 63 AINNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTTC 117
++N + DR+ AC+CIK + I G++ + + +CG P+ + PST C
Sbjct: 250 SLNAAYKSAEDRRGACQCIKDRAACIPGIDYTRVNQVGPRCGSKCPFKVYPSTNC 414
>AI938525 similar to SP|Q43304|NLTD_ Nonspecific lipid-transfer protein D
precursor (LTP D) (Wax-associated protein 9D).
[Cauliflower], partial (35%)
Length = 372
Score = 71.2 bits (173), Expect = 1e-13
Identities = 40/78 (51%), Positives = 52/78 (66%), Gaps = 1/78 (1%)
Frame = +2
Query: 6 VLVKVTCLTMMMCMVLGLPQTLDALSCLQVETKLMPCVPYVTGNGGYV-PQPCCDGVKAI 64
+LVK + L ++MCMVLG+P ALSC V+ + PC+ G GG V P PCC+GV+ +
Sbjct: 44 LLVKDSSL-VVMCMVLGMPLADAALSCP*VQFIVAPCLGD*RGLGGGVFPLPCCNGVRTL 220
Query: 65 NNQAVTKSDRQAACRCIK 82
NN A T DRQ ACRC+K
Sbjct: 221 NN*AKTTPDRQGACRCLK 274
>TC218866 weakly similar to UP|NLTP_CICAR (O23758) Nonspecific lipid-transfer
protein precursor (LTP), partial (83%)
Length = 814
Score = 68.9 bits (167), Expect = 5e-13
Identities = 39/115 (33%), Positives = 62/115 (53%), Gaps = 2/115 (1%)
Frame = +1
Query: 5 MVLVKVTCLTMMMCMVLGLPQTLDA-LSCLQVETKLMPCVPYVTGNGGYVPQPCCDGVKA 63
M +KV + ++ MV+ A ++C +V + PC+ Y+ GG CC+GV+
Sbjct: 76 MASLKVAFVVAVLYMVVVSAHMAHARINCGRVAVAVSPCLGYLR-RGGRPQARCCNGVRN 252
Query: 64 INNQAVTKSDRQAACRCIKTATSAI-HGLNMDILAGLPSKCGVHLPYTLGPSTTC 117
++ A T DR+ AC C+KT + G+N + A LP KC V++PY + ST C
Sbjct: 253 LHALAKTTVDRRTACNCLKTFARGLGRGVNANNAAALPRKCRVNIPYKISISTNC 417
>TC222645 similar to PRF|2115353A.0|1098265|2115353A lipid transfer protein.
{Hordeum vulgare;} , partial (27%)
Length = 487
Score = 68.2 bits (165), Expect = 9e-13
Identities = 28/75 (37%), Positives = 42/75 (55%)
Frame = +2
Query: 45 YVTGNGGYVPQPCCDGVKAINNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCG 104
Y+ G P CC G KA+ + A T D++AAC CIK+ + +I+ +N + LP CG
Sbjct: 8 YLVSGSGQPPAACCSGAKALASAATTSEDKKAACNCIKSTSKSIN-INSQLAQALPGNCG 184
Query: 105 VHLPYTLGPSTTCEK 119
+ LP + P+ C K
Sbjct: 185 ITLPVAISPNADCSK 229
>AI431156 similar to PIR|T12079|T120 non-specific lipid transfer protein
LTP-24 drought and ABA induced - kidney bean, partial
(51%)
Length = 196
Score = 67.4 bits (163), Expect = 2e-12
Identities = 31/61 (50%), Positives = 38/61 (61%)
Frame = -1
Query: 57 CCDGVKAINNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTLGPSTT 116
CC VK+IN A T +DRQ RC+K+A S I G+ I A LP KCGV +PY + ST
Sbjct: 193 CCIEVKSINAAAKTAADRQTESRCLKSAASQISGVEATIAASLPGKCGVSIPYKISTSTN 14
Query: 117 C 117
C
Sbjct: 13 C 11
>TC219531
Length = 576
Score = 67.0 bits (162), Expect = 2e-12
Identities = 35/110 (31%), Positives = 58/110 (51%), Gaps = 3/110 (2%)
Frame = +1
Query: 5 MVLVKVTCLTMMMCMVLGLPQTLDA---LSCLQVETKLMPCVPYVTGNGGYVPQPCCDGV 61
M V V LT+ + +VL + A +C + ++ + PC+ Y+T CC+GV
Sbjct: 112 MKKVFVAFLTLQVVLVLTIIAAEPAGKGYNCEKAKSSVTPCIKYLTSKVDTPSAACCNGV 291
Query: 62 KAINNQAVTKSDRQAACRCIKTATSAIHGLNMDILAGLPSKCGVHLPYTL 111
K + + A TK ++ AAC+C+K T+ I L D LP +CGV + + +
Sbjct: 292 KEVKSSAPTKDEKIAACQCLKEVTTHIPNLKEDRATALPKQCGVDVGFLI 441
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.135 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,891,969
Number of Sequences: 63676
Number of extensions: 95340
Number of successful extensions: 508
Number of sequences better than 10.0: 99
Number of HSP's better than 10.0 without gapping: 483
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 486
length of query: 125
length of database: 12,639,632
effective HSP length: 86
effective length of query: 39
effective length of database: 7,163,496
effective search space: 279376344
effective search space used: 279376344
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 53 (25.0 bits)
Medicago: description of AC147537.12


