

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147536.6 + phase: 0
(281 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
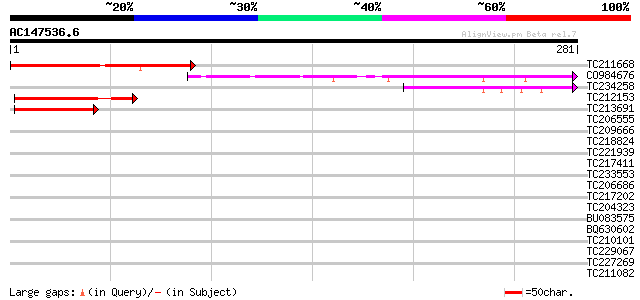
TC211668 similar to GB|AAM52242.1|21464575|AY120699 At2g39560/F1... 159 1e-39
CO984676 137 7e-33
TC234258 67 7e-12
TC212153 similar to GB|AAM52242.1|21464575|AY120699 At2g39560/F1... 64 6e-11
TC213691 similar to GB|AAM52242.1|21464575|AY120699 At2g39560/F1... 55 4e-08
TC206555 similar to UP|U315_ARATH (Q8LFJ5) UPF0315 protein At1g2... 30 0.94
TC209666 GB|AAB09252.1|1199563|SOYP34A 34 kDa maturing seed vacu... 30 1.6
TC218824 similar to GB|AAO39922.1|28372880|BT003694 At5g52960 {A... 30 1.6
TC221939 similar to UP|Q8GSM9 (Q8GSM9) Squalene monooxygenase 2 ... 29 2.7
TC217411 similar to UP|Q8L7S8 (Q8L7S8) At5g26740, partial (26%) 28 3.6
TC233553 similar to PIR|D69783|D69783 transcription regulator Ma... 28 3.6
TC206686 similar to UP|Q9SCP7 (Q9SCP7) Caffeic acid O-methyltran... 28 6.1
TC217202 homologue to UP|Q8W0W5 (Q8W0W5) Repressor protein, comp... 28 6.1
TC204323 similar to UP|NTF2_ARATH (Q9C7F5) Nuclear transport fac... 28 6.1
BU083575 weakly similar to GP|14279668|g verticillium wilt disea... 28 6.1
BQ630602 28 6.1
TC210101 27 7.9
TC229067 27 7.9
TC227269 similar to UP|Q8VZD9 (Q8VZD9) AT5g53140/MFH8_8, partial... 27 7.9
TC211082 27 7.9
>TC211668 similar to GB|AAM52242.1|21464575|AY120699 At2g39560/F12L6.22
{Arabidopsis thaliana;} , partial (17%)
Length = 819
Score = 159 bits (402), Expect = 1e-39
Identities = 77/93 (82%), Positives = 84/93 (89%), Gaps = 1/93 (1%)
Frame = -1
Query: 1 MKSLSSVGLGLSIVFGCLLLALVAEVYYLLWWKKKIIKREIENDYGNGNPLKELFYMFCW 60
MKSLSSVGLGLSI+FGCLLLALVAEVYYLLWWKK+I REIEN+Y GNP+KELFYMFCW
Sbjct: 273 MKSLSSVGLGLSIIFGCLLLALVAEVYYLLWWKKRITNREIENNY--GNPVKELFYMFCW 100
Query: 61 KRP-STSLRQTGFTPPELCSSMRINDNFVHDPQ 92
KRP S+SLRQT FTPPELCSSMRI D VH+P+
Sbjct: 99 KRPSSSSLRQTSFTPPELCSSMRIGDTLVHNPE 1
>CO984676
Length = 744
Score = 137 bits (344), Expect = 7e-33
Identities = 91/224 (40%), Positives = 128/224 (56%), Gaps = 31/224 (13%)
Frame = -2
Query: 89 HDPQVQTSKEFLFKPYGEDVIDADYMMQHHHDGLLGHPRFLFTIVEESKEDLESEDGKCG 148
H+P ++ + L K +GE+ +M+ H+ L G PRFLF I EE+KEDLESED +
Sbjct: 719 HEPDMEL--DLLHKXFGEESTVESELMRLHN--LAGPPRFLFPIKEETKEDLESED-RSR 555
Query: 149 KDSRGRSLGDL-LDVETPYLTPIASPHFFTPMNCSPYCS-------PYNQHGFNPLFEST 200
K SR RSL DL L ++TP+LTP+AS +P+ C CS Y GFNPLFES+
Sbjct: 554 KGSRTRSLSDLMLTIDTPFLTPVAS----SPLKC---CSLPVDPLHSYKHQGFNPLFESS 396
Query: 201 TDAEFNRLKSSPPPKFKFLQEAEEKLRRKMQDD----------------------NKGIN 238
T+ ++NR +SSPPPKFKF+++AEEKL RK+ ++ N+ +
Sbjct: 395 TELDYNRFRSSPPPKFKFMRDAEEKLHRKLMEEAARRKAVDNVQECCGLIKDSCSNETLA 216
Query: 239 GNEVDNSLITIIVDKK-YEREVNHHQCHLQQYHSSTSQVLPLAS 281
+ D S + I + K E Q +L Q+ S +SQ+LPLAS
Sbjct: 215 TDFSDGSFLKFIQNNKDTTTERKQLQQYLPQFPSGSSQILPLAS 84
>TC234258
Length = 623
Score = 67.4 bits (163), Expect = 7e-12
Identities = 41/102 (40%), Positives = 61/102 (59%), Gaps = 16/102 (15%)
Frame = +1
Query: 196 LFESTTDAEFNRLKSSPPPKFKFLQEAEEKLRRKMQDD--NKGINGNEV------DNSLI 247
LFES+ + E+NR +SSPPPKFKF+++AEEKL RK+ ++ K ++ N V D S
Sbjct: 10 LFESSVELEYNRFRSSPPPKFKFMRDAEEKLHRKLMEEARRKAVDNNNVQECGVKDCSNE 189
Query: 248 TIIVD------KKYEREVNHH--QCHLQQYHSSTSQVLPLAS 281
T+ D K+ + H Q ++ Q+ S + Q+LPLAS
Sbjct: 190 TLATDFRDGSFLKFIQTTEHKQPQQYVPQFPSGSFQILPLAS 315
>TC212153 similar to GB|AAM52242.1|21464575|AY120699 At2g39560/F12L6.22
{Arabidopsis thaliana;} , partial (12%)
Length = 655
Score = 64.3 bits (155), Expect = 6e-11
Identities = 30/61 (49%), Positives = 38/61 (62%)
Frame = +1
Query: 3 SLSSVGLGLSIVFGCLLLALVAEVYYLLWWKKKIIKREIENDYGNGNPLKELFYMFCWKR 62
S + + LS+VFGCLLLAL A++YY LWWKK+ + +IE D K FY CWK
Sbjct: 397 SFGGLSIALSLVFGCLLLALCAQLYYFLWWKKRRTQMDIEMD------AKGAFYWGCWKT 558
Query: 63 P 63
P
Sbjct: 559 P 561
>TC213691 similar to GB|AAM52242.1|21464575|AY120699 At2g39560/F12L6.22
{Arabidopsis thaliana;} , partial (12%)
Length = 638
Score = 55.1 bits (131), Expect = 4e-08
Identities = 23/42 (54%), Positives = 31/42 (73%)
Frame = +1
Query: 3 SLSSVGLGLSIVFGCLLLALVAEVYYLLWWKKKIIKREIEND 44
S + + LS+VFGCLLLAL A++YY LWWKK+ + +IE D
Sbjct: 502 SFGGLSIALSLVFGCLLLALCAQLYYFLWWKKRRTQMDIEMD 627
>TC206555 similar to UP|U315_ARATH (Q8LFJ5) UPF0315 protein At1g22270,
partial (90%)
Length = 621
Score = 30.4 bits (67), Expect = 0.94
Identities = 19/58 (32%), Positives = 32/58 (54%), Gaps = 2/58 (3%)
Frame = +3
Query: 34 KKIIKREIENDYGNGNPLKELFYMFCWKRPSTSLRQTGFT--PPELCSSMRINDNFVH 89
+K++++ +E NG LK++F WK + R G+T P E SSM +D F++
Sbjct: 300 EKVVEKTVEM---NGEFLKKMFEKIDWKAFVDASRAMGYTELPEEADSSMLDSDEFLN 464
>TC209666 GB|AAB09252.1|1199563|SOYP34A 34 kDa maturing seed vacuolar thiol
protease precursor {Glycine max;} , partial (8%)
Length = 572
Score = 29.6 bits (65), Expect = 1.6
Identities = 21/57 (36%), Positives = 28/57 (48%)
Frame = +3
Query: 158 DLLDVETPYLTPIASPHFFTPMNCSPYCSPYNQHGFNPLFESTTDAEFNRLKSSPPP 214
DLL +T +P S H P C+P S + F LF S + ++ L SSPPP
Sbjct: 207 DLLHPQTSS-SPSQSRHANPPRLCAPLMSSSTRASFC-LFTSPNASPWSALSSSPPP 371
>TC218824 similar to GB|AAO39922.1|28372880|BT003694 At5g52960 {Arabidopsis
thaliana;} , partial (57%)
Length = 831
Score = 29.6 bits (65), Expect = 1.6
Identities = 23/66 (34%), Positives = 30/66 (44%)
Frame = +2
Query: 165 PYLTPIASPHFFTPMNCSPYCSPYNQHGFNPLFESTTDAEFNRLKSSPPPKFKFLQEAEE 224
PY P S HFFTP +P+ SP+ F P + F L +SP + Q
Sbjct: 176 PY--PHKSSHFFTPFKPNPFPSPFLI*EFKPHHHT-----FPHLSASP----RRAQRRRP 322
Query: 225 KLRRKM 230
+ RRKM
Sbjct: 323 RPRRKM 340
>TC221939 similar to UP|Q8GSM9 (Q8GSM9) Squalene monooxygenase 2 , partial
(7%)
Length = 695
Score = 28.9 bits (63), Expect = 2.7
Identities = 14/42 (33%), Positives = 24/42 (56%), Gaps = 1/42 (2%)
Frame = +3
Query: 199 STTDAEFNRLKSSPPPKFKFLQEAEEKLRRKMQDDNKG-ING 239
S E NR ++ PP + ++ K+RR+ DD++G +NG
Sbjct: 510 SGRSTESNRERTDPPFRLNKTEKPRRKMRRRNDDDDEGEMNG 635
>TC217411 similar to UP|Q8L7S8 (Q8L7S8) At5g26740, partial (26%)
Length = 1275
Score = 28.5 bits (62), Expect = 3.6
Identities = 23/67 (34%), Positives = 30/67 (44%), Gaps = 14/67 (20%)
Frame = -2
Query: 32 WKKKIIKREIENDYGNGN-----PLKELFYMFCWKRPSTSLRQTGFTPPELC-------- 78
W+ I++R D GNG P+K LF F WK + SL P +C
Sbjct: 500 WRPIILQRR*LGDLGNGVSRWYVPIK*LFSYFLWKIKNGSLN------PFICN*MYFSNF 339
Query: 79 -SSMRIN 84
SS+RIN
Sbjct: 338 ISSLRIN 318
>TC233553 similar to PIR|D69783|D69783 transcription regulator MarR family
homolog ydgJ - Bacillus subtilis {Bacillus subtilis;} ,
partial (88%)
Length = 697
Score = 28.5 bits (62), Expect = 3.6
Identities = 14/34 (41%), Positives = 18/34 (52%)
Frame = -1
Query: 54 LFYMFCWKRPSTSLRQTGFTPPELCSSMRINDNF 87
+F MFCW R STS+R + S IN +F
Sbjct: 487 IFSMFCWIRMSTSVRSLRSSSLTFSSISEINRSF 386
>TC206686 similar to UP|Q9SCP7 (Q9SCP7) Caffeic acid O-methyltransferase-like
protein (AT3g53140/T4D2_70), partial (89%)
Length = 1298
Score = 27.7 bits (60), Expect = 6.1
Identities = 13/37 (35%), Positives = 21/37 (56%), Gaps = 1/37 (2%)
Frame = +1
Query: 111 ADYMMQHHHDGLL-GHPRFLFTIVEESKEDLESEDGK 146
A Y++QHH D L+ P +V+ +KE E +G+
Sbjct: 337 AHYVLQHHQDALMRAWPMVHEAVVDPTKEPFERANGE 447
>TC217202 homologue to UP|Q8W0W5 (Q8W0W5) Repressor protein, complete
Length = 852
Score = 27.7 bits (60), Expect = 6.1
Identities = 28/99 (28%), Positives = 38/99 (38%), Gaps = 15/99 (15%)
Frame = -3
Query: 116 QHHHDGLLGHPRFLFTIVEESKEDLESEDGKC---GKDSRGRSL--GDLLDV-------- 162
+H H G RFL+TI D S+ G C KD + L G L
Sbjct: 697 EHIHIGRQTSERFLYTIELIESTDRRSQGGACLCFNKDPKVSYLNFGQHLAA*KQWLHHS 518
Query: 163 --ETPYLTPIASPHFFTPMNCSPYCSPYNQHGFNPLFES 199
P T A+ P + +CS Y+ H +PL +S
Sbjct: 517 S*HVPLQTSFAALLMLLPQSSQLHCSIYHHH--HPLLKS 407
>TC204323 similar to UP|NTF2_ARATH (Q9C7F5) Nuclear transport factor 2
(NTF-2), partial (98%)
Length = 812
Score = 27.7 bits (60), Expect = 6.1
Identities = 12/25 (48%), Positives = 18/25 (72%), Gaps = 1/25 (4%)
Frame = -1
Query: 59 CWKRP-STSLRQTGFTPPELCSSMR 82
CWKRP S++LR++ + PP+ S R
Sbjct: 218 CWKRPGSSALRRSAWNPPDTDSLNR 144
>BU083575 weakly similar to GP|14279668|g verticillium wilt disease
resistance protein {Lycopersicon esculentum}, partial
(9%)
Length = 392
Score = 27.7 bits (60), Expect = 6.1
Identities = 22/55 (40%), Positives = 25/55 (45%), Gaps = 7/55 (12%)
Frame = -3
Query: 47 NGNPLKELFYMFCWKRPSTSLRQTGF--TPP-ELCSSMRIN-DNFVHD---PQVQ 94
N L LF CW R LRQ GF TP LC + N DN H P++Q
Sbjct: 189 NS*ALPVLFLQNCWMRSQQMLRQLGF*QTPSVNLCL*VSPNPDNIYHQTKGPEIQ 25
>BQ630602
Length = 432
Score = 27.7 bits (60), Expect = 6.1
Identities = 12/24 (50%), Positives = 18/24 (75%)
Frame = -2
Query: 253 KKYEREVNHHQCHLQQYHSSTSQV 276
K+ EREV ++ CH Q+HSS +Q+
Sbjct: 77 KRKEREVPNNMCH-SQHHSSIAQI 9
>TC210101
Length = 571
Score = 27.3 bits (59), Expect = 7.9
Identities = 10/23 (43%), Positives = 12/23 (51%)
Frame = -3
Query: 165 PYLTPIASPHFFTPMNCSPYCSP 187
P P PH + CSP+CSP
Sbjct: 245 PSSAPSLDPHILSLPPCSPFCSP 177
>TC229067
Length = 1161
Score = 27.3 bits (59), Expect = 7.9
Identities = 12/39 (30%), Positives = 19/39 (47%)
Frame = +2
Query: 241 EVDNSLITIIVDKKYEREVNHHQCHLQQYHSSTSQVLPL 279
E+D+ L I K+ +R + H+ QYH S + L
Sbjct: 644 EIDDKLFLCIPQKELQRRYSKHESTSYQYHPSWDDIYGL 760
>TC227269 similar to UP|Q8VZD9 (Q8VZD9) AT5g53140/MFH8_8, partial (70%)
Length = 1398
Score = 27.3 bits (59), Expect = 7.9
Identities = 10/19 (52%), Positives = 14/19 (73%)
Frame = +2
Query: 111 ADYMMQHHHDGLLGHPRFL 129
A+Y+ +H D LL HP+FL
Sbjct: 458 AEYLKEHLFDNLLKHPKFL 514
>TC211082
Length = 518
Score = 27.3 bits (59), Expect = 7.9
Identities = 17/45 (37%), Positives = 23/45 (50%), Gaps = 6/45 (13%)
Frame = -1
Query: 11 LSIVFGCLLLALVAEVYYLLWWKKKIIKREIE------NDYGNGN 49
L + F L+LALV VY+LL W + + E+ DY N N
Sbjct: 158 LKVDFYQLILALVCLVYFLLLW*VESVTSELHANT*KFTDYSN*N 24
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.137 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,645,283
Number of Sequences: 63676
Number of extensions: 242651
Number of successful extensions: 1676
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 1586
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1664
length of query: 281
length of database: 12,639,632
effective HSP length: 96
effective length of query: 185
effective length of database: 6,526,736
effective search space: 1207446160
effective search space used: 1207446160
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC147536.6


