

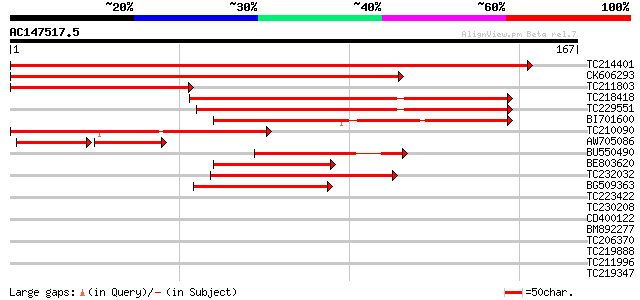
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147517.5 + phase: 0
(167 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC214401 weakly similar to UP|Q949G3 (Q949G3) ABC1 protein, part... 224 1e-59
CK606293 195 1e-50
TC211803 similar to UP|Q76CU2 (Q76CU2) PDR-type ABC transporter ... 74 2e-14
TC218418 homologue to UP|Q6X4V5 (Q6X4V5) ABC transporter, partia... 71 2e-13
TC229551 weakly similar to UP|Q6X4V5 (Q6X4V5) ABC transporter, p... 70 5e-13
BI701600 63 7e-11
TC210090 similar to UP|Q9LFH0 (Q9LFH0) PDR9 ABC transporter, par... 56 9e-09
AW705086 43 2e-07
BU550490 48 2e-06
BE803620 homologue to GP|13605839|gb| At2g01320/F10A8.20 {Arabid... 45 2e-05
TC232032 weakly similar to UP|Q6K376 (Q6K376) ABC transporter-li... 44 5e-05
BG509363 similar to GP|14331118|e ABC1 protein {Nicotiana plumba... 40 5e-04
TC223422 39 0.001
TC230208 similar to UP|Q6YUV9 (Q6YUV9) Transitional endoplasmic ... 38 0.003
CD400122 38 0.003
BM892277 38 0.003
TC206370 similar to UP|Q940D1 (Q940D1) At1g64110/F22C12_22, part... 37 0.004
TC219888 similar to UP|Q944N4 (Q944N4) Tobacco mosaic virus heli... 35 0.022
TC211996 similar to UP|Q940D1 (Q940D1) At1g64110/F22C12_22, part... 34 0.029
TC219347 similar to UP|Q9LF78 (Q9LF78) Mitochondrial half-ABC tr... 33 0.083
>TC214401 weakly similar to UP|Q949G3 (Q949G3) ABC1 protein, partial (18%)
Length = 1177
Score = 224 bits (571), Expect = 1e-59
Identities = 108/154 (70%), Positives = 127/154 (82%)
Frame = +1
Query: 1 MDRVGIDLPTIEVRFEHLNIEAEAHVGSISLPTFTNFMVNIVESLLNSLHVLPSRKQRLN 60
MDRVG+D+PTIEVRFEH+N+EA+ +VG +LP+ NF N++E LN LH++PS K+ L
Sbjct: 583 MDRVGLDIPTIEVRFEHINVEAQVYVGGRALPSMLNFFANVIEGFLNYLHIIPSPKKPLR 762
Query: 61 ILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMSEFVPQR 120
IL++VSGIIKP RMTLLLGPP SGKTTLLLALAGKLD L SGRVTYNGH + EFVPQR
Sbjct: 763 ILQNVSGIIKPRRMTLLLGPPGSGKTTLLLALAGKLDKDLNHSGRVTYNGHGLEEFVPQR 942
Query: 121 TAAYVDQNDLHIGELTVRETLAFSARVQGVGPQY 154
T+AY+ Q D HIGE+TVRETLAFSAR QGVG Y
Sbjct: 943 TSAYISQYDNHIGEMTVRETLAFSARCQGVGQNY 1044
>CK606293
Length = 782
Score = 195 bits (495), Expect = 1e-50
Identities = 97/116 (83%), Positives = 108/116 (92%)
Frame = -3
Query: 1 MDRVGIDLPTIEVRFEHLNIEAEAHVGSISLPTFTNFMVNIVESLLNSLHVLPSRKQRLN 60
+DRVGIDLPTIEVRFE+L+IEAEA G+ +LPTFTNF+VNI+E LLNSLHVLP+RKQ LN
Sbjct: 348 IDRVGIDLPTIEVRFENLSIEAEARAGTRALPTFTNFIVNILEGLLNSLHVLPNRKQHLN 169
Query: 61 ILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMSEF 116
IL+DVSGIIKP RMTLLLGPPSSGKTTLLLALAGKLDPK KFSG+VTYNGH M+EF
Sbjct: 168 ILEDVSGIIKPGRMTLLLGPPSSGKTTLLLALAGKLDPKKKFSGKVTYNGHGMNEF 1
>TC211803 similar to UP|Q76CU2 (Q76CU2) PDR-type ABC transporter 1, partial
(6%)
Length = 255
Score = 74.3 bits (181), Expect = 2e-14
Identities = 34/54 (62%), Positives = 43/54 (78%)
Frame = +1
Query: 1 MDRVGIDLPTIEVRFEHLNIEAEAHVGSISLPTFTNFMVNIVESLLNSLHVLPS 54
+DRVG+D+PTIEVR+EHLNIEAEA VGS +LP+F N + N+VE N LH+ S
Sbjct: 94 IDRVGLDIPTIEVRYEHLNIEAEAFVGSRALPSFINSVTNVVEGFFNLLHISTS 255
>TC218418 homologue to UP|Q6X4V5 (Q6X4V5) ABC transporter, partial (49%)
Length = 1220
Score = 71.2 bits (173), Expect = 2e-13
Identities = 40/95 (42%), Positives = 59/95 (62%)
Frame = +1
Query: 54 SRKQRLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEM 113
S + N+L+ ++G +P T L+GP SGK+TLL AL+ +L SG + NG +
Sbjct: 277 SNGETQNVLEGLTGYAEPGTFTALMGPSGSGKSTLLDALSSRLAANAFLSGTILLNGRKA 456
Query: 114 SEFVPQRTAAYVDQNDLHIGELTVRETLAFSARVQ 148
+ TAAYV Q+D IG LTVRET+++SAR++
Sbjct: 457 K--LSFGTAAYVTQDDNLIGTLTVRETISYSARLR 555
>TC229551 weakly similar to UP|Q6X4V5 (Q6X4V5) ABC transporter, partial (34%)
Length = 1436
Score = 70.1 bits (170), Expect = 5e-13
Identities = 38/93 (40%), Positives = 57/93 (60%)
Frame = +3
Query: 56 KQRLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMSE 115
K R IL ++G +P R+ ++GP SGK+TLL ALAG+L +K +G++ NGH+
Sbjct: 60 KNRKLILHGLTGYAQPGRLLAIIGPSGSGKSTLLDALAGRLTSNIKQTGKILINGHKQE- 236
Query: 116 FVPQRTAAYVDQNDLHIGELTVRETLAFSARVQ 148
+ T+ YV Q+D + LT ETL +SA +Q
Sbjct: 237 -LAYGTSGYVTQDDAMLSCLTAGETLYYSAMLQ 332
>BI701600
Length = 426
Score = 62.8 bits (151), Expect = 7e-11
Identities = 40/89 (44%), Positives = 58/89 (64%), Gaps = 1/89 (1%)
Frame = +3
Query: 61 ILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKL-DPKLKFSGRVTYNGHEMSEFVPQ 119
ILK V+GI +P + +LGP SGK+TLL ALAG+L P L +G + N ++++ V +
Sbjct: 72 ILKGVTGIAQPGEILAVLGPSGSGKSTLLHALAGRLHGPGL--TGTILANSSKLTKPVLR 245
Query: 120 RTAAYVDQNDLHIGELTVRETLAFSARVQ 148
RT +V Q+D+ LTVRETL F A ++
Sbjct: 246 RT-GFVTQDDILYPHLTVRETLVFCAMLR 329
>TC210090 similar to UP|Q9LFH0 (Q9LFH0) PDR9 ABC transporter, partial (12%)
Length = 1189
Score = 55.8 bits (133), Expect = 9e-09
Identities = 30/78 (38%), Positives = 51/78 (64%), Gaps = 1/78 (1%)
Frame = +3
Query: 1 MDRVGIDLPTIEVRFEHLNIEAEAH-VGSISLPTFTNFMVNIVESLLNSLHVLPSRKQRL 59
+D+VGI+LPT+E+R+++L++EAE V +PT N + + L VL S+ ++
Sbjct: 390 IDKVGINLPTVELRYQNLSVEAECKIVQGKPIPTLWNTLKEWIFD-TTKLSVLKSQNSKI 566
Query: 60 NILKDVSGIIKPSRMTLL 77
+I+K+ +GIIKP R +L
Sbjct: 567 SIIKNDNGIIKPGRYAIL 620
>AW705086
Length = 427
Score = 43.1 bits (100), Expect(2) = 2e-07
Identities = 18/22 (81%), Positives = 22/22 (99%)
Frame = -3
Query: 3 RVGIDLPTIEVRFEHLNIEAEA 24
RVGID+PTIEVR++HLN+EAEA
Sbjct: 242 RVGIDIPTIEVRYKHLNVEAEA 177
Score = 28.1 bits (61), Expect(2) = 2e-07
Identities = 13/21 (61%), Positives = 15/21 (70%)
Frame = -2
Query: 26 VGSISLPTFTNFMVNIVESLL 46
VGS +LPTF NF+ N VE L
Sbjct: 174 VGSRALPTFVNFLTNKVEVCL 112
>BU550490
Length = 616
Score = 48.1 bits (113), Expect = 2e-06
Identities = 26/45 (57%), Positives = 31/45 (68%)
Frame = +2
Query: 73 RMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMSEFV 117
R+TLLLGPP SGKTTLL ALAGKLD L+ GH ++ F+
Sbjct: 71 RLTLLLGPPRSGKTTLLQALAGKLDRDLRV-------GHPLAPFL 184
>BE803620 homologue to GP|13605839|gb| At2g01320/F10A8.20 {Arabidopsis
thaliana}, partial (8%)
Length = 413
Score = 44.7 bits (104), Expect = 2e-05
Identities = 21/36 (58%), Positives = 27/36 (74%)
Frame = +3
Query: 61 ILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKL 96
+LK+VSG KP R+ ++GP SGKTTLL LAG+L
Sbjct: 294 LLKNVSGEAKPGRLLAIMGPSGSGKTTLLNVLAGQL 401
>TC232032 weakly similar to UP|Q6K376 (Q6K376) ABC transporter-like protein,
partial (11%)
Length = 799
Score = 43.5 bits (101), Expect = 5e-05
Identities = 19/55 (34%), Positives = 34/55 (61%)
Frame = +2
Query: 60 NILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMS 114
+IL+ ++G KP ++ ++GP GK+TLL LAG+L + +G + NG + +
Sbjct: 617 SILEGLTGYAKPGQLLAIMGPSGCGKSTLLDTLAGRLGSNTRQTGEILINGKKQA 781
>BG509363 similar to GP|14331118|e ABC1 protein {Nicotiana plumbaginifolia},
partial (8%)
Length = 455
Score = 40.0 bits (92), Expect = 5e-04
Identities = 19/41 (46%), Positives = 28/41 (67%)
Frame = +1
Query: 55 RKQRLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGK 95
++ RL +LK VSG +P +T L+G +GKTTL+ LAG+
Sbjct: 313 QEDRLVLLKGVSGAFRPGVLTALMGVSGAGKTTLMDVLAGR 435
>TC223422
Length = 426
Score = 38.9 bits (89), Expect = 0.001
Identities = 23/44 (52%), Positives = 28/44 (63%), Gaps = 2/44 (4%)
Frame = +2
Query: 76 LLLGPPSSGKTTLLLALAGKLDPKLK--FSGRVTYNGHEMSEFV 117
+ LGP SSGK LLLALAG+LDP LK SG V ++S +
Sbjct: 158 IALGPTSSGKNILLLALAGELDP*LKTFTSGVVEKKKRQISSMI 289
>TC230208 similar to UP|Q6YUV9 (Q6YUV9) Transitional endoplasmic reticulum
ATPase-like, partial (45%)
Length = 642
Score = 37.7 bits (86), Expect = 0.003
Identities = 27/79 (34%), Positives = 37/79 (46%)
Frame = +1
Query: 15 FEHLNIEAEAHVGSISLPTFTNFMVNIVESLLNSLHVLPSRKQRLNILKDVSGIIKPSRM 74
FE I A G I + + V+ LN L +LP R+ L +++P +
Sbjct: 262 FESNFISAVVPPGEIGVKFDDIGALEDVKKALNELVILPMRRPELF---SRGNLLRPCKG 432
Query: 75 TLLLGPPSSGKTTLLLALA 93
LL GPP +GKT L ALA
Sbjct: 433 ILLFGPPGTGKTLLAKALA 489
>CD400122
Length = 585
Score = 37.7 bits (86), Expect = 0.003
Identities = 18/39 (46%), Positives = 25/39 (63%)
Frame = -1
Query: 58 RLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKL 96
R IL+ + G KP ++ ++GP GK+TLL ALAG L
Sbjct: 243 RKPILQGLKGYAKPGKLLAIMGPSGCGKSTLLDALAGNL 127
>BM892277
Length = 421
Score = 37.7 bits (86), Expect = 0.003
Identities = 23/66 (34%), Positives = 38/66 (56%)
Frame = +2
Query: 60 NILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMSEFVPQ 119
++LK+VS KP + ++GP +GK++LL LAGK P+ SG V N + + +
Sbjct: 233 HVLKNVSFQAKPWEILAIVGPSGAGKSSLLEILAGKHSPQ---SGTVFLNHKPVDKAQFR 403
Query: 120 RTAAYV 125
+ + YV
Sbjct: 404 KLSGYV 421
>TC206370 similar to UP|Q940D1 (Q940D1) At1g64110/F22C12_22, partial (38%)
Length = 1502
Score = 37.0 bits (84), Expect = 0.004
Identities = 22/54 (40%), Positives = 32/54 (58%)
Frame = +2
Query: 42 VESLLNSLHVLPSRKQRLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGK 95
++ L L +LP R R ++ K G++KP R LL GPP +GKT L A+A +
Sbjct: 86 IKESLQELVMLPLR--RPDLFK--GGLLKPCRGILLFGPPGTGKTMLAKAIANE 235
>TC219888 similar to UP|Q944N4 (Q944N4) Tobacco mosaic virus helicase
domain-binding protein (Fragment), partial (50%)
Length = 1119
Score = 34.7 bits (78), Expect = 0.022
Identities = 18/50 (36%), Positives = 31/50 (62%)
Frame = +3
Query: 46 LNSLHVLPSRKQRLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGK 95
L + +LP++++ L +G+ +P+R LL GPP +GKT L A+A +
Sbjct: 21 LMEMVILPTKRRDL-----FTGLRRPARGLLLFGPPGNGKTMLAKAVASE 155
>TC211996 similar to UP|Q940D1 (Q940D1) At1g64110/F22C12_22, partial (17%)
Length = 580
Score = 34.3 bits (77), Expect = 0.029
Identities = 20/50 (40%), Positives = 28/50 (56%)
Frame = +3
Query: 46 LNSLHVLPSRKQRLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGK 95
L L +LP R+ L G++KP + LL GPP +GKT L A+A +
Sbjct: 417 LQELVMLPLRRPDLFR----GGLLKPCKGILLFGPPGTGKTMLAKAIASE 554
>TC219347 similar to UP|Q9LF78 (Q9LF78) Mitochondrial half-ABC transporter,
partial (36%)
Length = 1085
Score = 32.7 bits (73), Expect = 0.083
Identities = 26/101 (25%), Positives = 43/101 (41%), Gaps = 13/101 (12%)
Frame = +1
Query: 61 ILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMSE----- 115
IL +S ++ + ++G SGK+T+L L DP SG + + + E
Sbjct: 97 ILDGISFVVPAGKSVAIVGTSGSGKSTILRLLFRFFDP---HSGSIKIDDQNIREVTLES 267
Query: 116 ------FVPQRTAAYVDQ--NDLHIGELTVRETLAFSARVQ 148
VPQ T + D +++H G L+ E + A Q
Sbjct: 268 LRKSIGVVPQDTVLFNDTIFHNIHYGRLSATEEEVYEAAQQ 390
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.138 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,433,468
Number of Sequences: 63676
Number of extensions: 76962
Number of successful extensions: 454
Number of sequences better than 10.0: 97
Number of HSP's better than 10.0 without gapping: 450
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 452
length of query: 167
length of database: 12,639,632
effective HSP length: 90
effective length of query: 77
effective length of database: 6,908,792
effective search space: 531976984
effective search space used: 531976984
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 55 (25.8 bits)
Medicago: description of AC147517.5


