

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
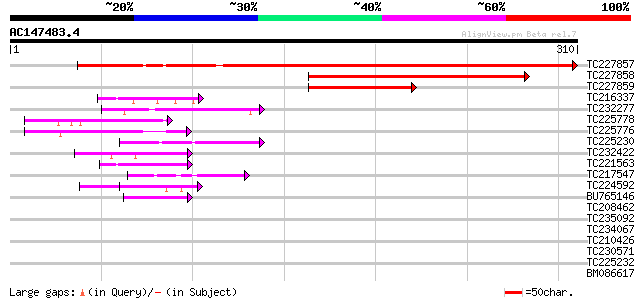
Query= AC147483.4 + phase: 0
(310 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC227857 weakly similar to UP|Q8LA02 (Q8LA02) Contains similarit... 365 e-101
TC227858 similar to UP|MTR4_YEAST (P47047) ATP-dependent RNA hel... 201 3e-52
TC227859 104 6e-23
TC216337 similar to UP|Q7XJ13 (Q7XJ13) Chloroplast nucleoid DNA-... 47 1e-05
TC232277 similar to UP|Q756S8 (Q756S8) AER176Wp, partial (7%) 47 1e-05
TC225778 similar to UP|Q93YH8 (Q93YH8) HMG I/Y like protein, par... 45 4e-05
TC225776 similar to UP|Q93YH8 (Q93YH8) HMG I/Y like protein, par... 45 4e-05
TC225230 similar to UP|Q9SZZ1 (Q9SZZ1) Fibrillarin-like protein ... 45 5e-05
TC232422 weakly similar to UP|Q41645 (Q41645) Extensin (Fragment... 44 1e-04
TC221563 similar to UP|O23702 (O23702) Dehydrogenase, partial (21%) 44 1e-04
TC217547 weakly similar to UP|Q61869 (Q61869) Keratin 2 epidermi... 42 4e-04
TC224592 similar to UP|Q93VA8 (Q93VA8) At1g76010/T4O12_22, parti... 42 5e-04
BU765146 similar to GP|21322711|e pherophorin-dz1 protein {Volvo... 41 8e-04
TC208462 homologue to UP|LSM4_TOBAC (Q43582) Probable U6 snRNA-a... 40 0.001
TC235092 similar to UP|O91332 (O91332) EBNA-1, partial (5%) 40 0.001
TC234067 weakly similar to UP|Q9SBM1 (Q9SBM1) Hydroxyproline-ric... 39 0.002
TC210426 similar to UP|Q9FK53 (Q9FK53) Similarity to GAR1 protei... 39 0.003
TC230571 similar to UP|Q41645 (Q41645) Extensin (Fragment), part... 39 0.003
TC225232 similar to UP|Q9SZZ1 (Q9SZZ1) Fibrillarin-like protein ... 39 0.003
BM086617 homologue to GP|9651941|gb receptor-like protein kinase... 38 0.005
>TC227857 weakly similar to UP|Q8LA02 (Q8LA02) Contains similarity to
RNA-binding protein from Arabidopsis thaliana gi|2129727
and contains RNA recognition PF|00076 domain, partial
(37%)
Length = 995
Score = 365 bits (936), Expect = e-101
Identities = 187/274 (68%), Positives = 220/274 (80%), Gaps = 1/274 (0%)
Frame = +2
Query: 38 RQTPADAESDNVPRRQPMNRFVRDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDG 97
+++P ++ D + +DDG +GRGR G GR RGRG RGRG GRG G
Sbjct: 2 KRSPIPSQEDATRNALKILSHGKDDGSDTGRGREYG-GRGGLDRGRG-RGRGRGRGRGMG 175
Query: 98 RGGF-KRYGDDRTSHQDIARSNADGLYVGDNADGEKLAKKLGPEIMDQITEAYEEIIERV 156
RG F +R D++ D A GLY GD+ADGEKLA+K+GPEIM+Q+TE +EE+ RV
Sbjct: 176 RGRFVERDVDEKVMDTD---DYATGLYAGDDADGEKLARKVGPEIMNQLTEGFEEMTSRV 346
Query: 157 LPSPLQDEYVEAMDINCAIEFEPEYAVEFDNPDIDEKEPIALRDALEKMKPFLMTYEGIR 216
LPSPL+DE+++A+DIN AIEFEPEY VEFDNPDIDEKEPI+LRDALEK KPFLM+YEGI+
Sbjct: 347 LPSPLEDEFLDALDINYAIEFEPEYLVEFDNPDIDEKEPISLRDALEKAKPFLMSYEGIQ 526
Query: 217 SQEEWEEVIEELMQRVPLLKKIVDHYSGPDRVTAKKQQEELERVAKTLPTSAPSSVKEFT 276
SQEEWEE++EE M RVPLLKKI+DHYSGPDRVTAKKQQEELERVAKTLP S PSSVK+FT
Sbjct: 527 SQEEWEEIMEETMARVPLLKKIIDHYSGPDRVTAKKQQEELERVAKTLPGSVPSSVKQFT 706
Query: 277 NRAVVSLQSNPGWGFDKKCQFMDKLVFEVSQHHK 310
NRAV+SLQSNPGWGFDKKC FMDKLV+EVSQH+K
Sbjct: 707 NRAVISLQSNPGWGFDKKCHFMDKLVWEVSQHYK 808
>TC227858 similar to UP|MTR4_YEAST (P47047) ATP-dependent RNA helicase DOB1
(mRNA transport regulator MTR4), partial (5%)
Length = 617
Score = 201 bits (512), Expect = 3e-52
Identities = 97/121 (80%), Positives = 111/121 (91%)
Frame = +3
Query: 164 EYVEAMDINCAIEFEPEYAVEFDNPDIDEKEPIALRDALEKMKPFLMTYEGIRSQEEWEE 223
E+++A+DIN AIEFEPEY VEFDNPDIDEKEPI LRDALEK KPFLM+YEGI+SQE WE+
Sbjct: 252 EFLDALDINYAIEFEPEYLVEFDNPDIDEKEPIPLRDALEKAKPFLMSYEGIQSQEVWEK 431
Query: 224 VIEELMQRVPLLKKIVDHYSGPDRVTAKKQQEELERVAKTLPTSAPSSVKEFTNRAVVSL 283
+EE M RVPLLKKIVDHYSGPDRVT KK+QEEL+RVAKTLP SAPSSV++FTNR+V+SL
Sbjct: 432 TMEETMARVPLLKKIVDHYSGPDRVTTKKRQEELDRVAKTLPESAPSSVEQFTNRSVISL 611
Query: 284 Q 284
Q
Sbjct: 612 Q 614
>TC227859
Length = 1031
Score = 104 bits (259), Expect = 6e-23
Identities = 48/59 (81%), Positives = 54/59 (91%)
Frame = +3
Query: 164 EYVEAMDINCAIEFEPEYAVEFDNPDIDEKEPIALRDALEKMKPFLMTYEGIRSQEEWE 222
E+++A+DIN AIEFEPEY VEFDNPDIDEKEPI LRDALEK KPFLM+YEGI+SQE WE
Sbjct: 615 EFLDALDINYAIEFEPEYLVEFDNPDIDEKEPIPLRDALEKAKPFLMSYEGIQSQEVWE 791
>TC216337 similar to UP|Q7XJ13 (Q7XJ13) Chloroplast nucleoid DNA-binding
protein-like protein, partial (21%)
Length = 1139
Score = 46.6 bits (109), Expect = 1e-05
Identities = 34/85 (40%), Positives = 43/85 (50%), Gaps = 27/85 (31%)
Frame = +3
Query: 49 VPRRQPMNRFVRDDGDGS------GRGRGRGRGRDVY------------ARGRGDRGRG- 89
V + +P+N + ++G+GS GRGRGRGRGR Y RG G RGRG
Sbjct: 528 VDQVKPLNEY-EEEGEGSPRMRGRGRGRGRGRGRGFYNGGMEYGDDWDGGRGYGGRGRGR 704
Query: 90 --GRGGRGDGRG------GFKRYGD 106
GR RG GRG G+ YG+
Sbjct: 705 ARGRAFRGRGRGYGVQPVGYYDYGE 779
Score = 39.3 bits (90), Expect = 0.002
Identities = 27/62 (43%), Positives = 27/62 (43%), Gaps = 23/62 (37%)
Frame = +3
Query: 61 DDGDGS----GRGRGRGRGRDVYARGRG-------------------DRGRGGRGGRGDG 97
DD DG GRGRGR RGR RGRG RGRG GRG G
Sbjct: 657 DDWDGGRGYGGRGRGRARGRAFRGRGRGYGVQPVGYYDYGEYDAQPPPRGRGRGRGRGRG 836
Query: 98 RG 99
RG
Sbjct: 837 RG 842
Score = 31.6 bits (70), Expect = 0.48
Identities = 13/15 (86%), Positives = 13/15 (86%)
Frame = +3
Query: 63 GDGSGRGRGRGRGRD 77
G G GRGRGRGRGRD
Sbjct: 804 GRGRGRGRGRGRGRD 848
Score = 28.9 bits (63), Expect = 3.1
Identities = 17/27 (62%), Positives = 17/27 (62%), Gaps = 1/27 (3%)
Frame = +3
Query: 62 DGDGSGRGRGRGRGRDVYARGRG-DRG 87
D RGRGRGRGR RGRG DRG
Sbjct: 783 DAQPPPRGRGRGRGR---GRGRGRDRG 854
Score = 27.3 bits (59), Expect = 9.0
Identities = 12/17 (70%), Positives = 12/17 (70%)
Frame = +3
Query: 60 RDDGDGSGRGRGRGRGR 76
R G G GRGRGRGR R
Sbjct: 801 RGRGRGRGRGRGRGRDR 851
>TC232277 similar to UP|Q756S8 (Q756S8) AER176Wp, partial (7%)
Length = 445
Score = 46.6 bits (109), Expect = 1e-05
Identities = 38/96 (39%), Positives = 46/96 (47%), Gaps = 7/96 (7%)
Frame = -3
Query: 51 RRQPMNRFVRD--DGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDR 108
RR P+ R R DG G+ RGR RGRGR +G R RGG GGR G G R G R
Sbjct: 392 RRSPLRRGGRRGRDGGGAWRGRRRGRGR---RSRQGRRRRGGEGGRA*GAPGACRRGRGR 222
Query: 109 TSH-QDIARSNADGLYVGDNADG----EKLAKKLGP 139
H +D R GL + + E L + +GP
Sbjct: 221 F*HRRDGFRGRRSGLETEKSTNSRVW*ESLLRLVGP 114
>TC225778 similar to UP|Q93YH8 (Q93YH8) HMG I/Y like protein, partial (12%)
Length = 417
Score = 45.1 bits (105), Expect = 4e-05
Identities = 34/91 (37%), Positives = 45/91 (49%), Gaps = 10/91 (10%)
Frame = +1
Query: 9 GGGRGRGKPLEEAAQEA--PQAPVVN------RHIRV--RQTPADAESDNVPRRQPMNRF 58
G R G+P + +Q P AP V+ H+ P DA++ PR + +
Sbjct: 4 GPKRRPGRPPKNQSQPTLIPFAPAVSAASVDTEHVAASAETAPVDADAALGPRARGRPKK 183
Query: 59 VRDDGDGSGRGRGRGRGRDVYARGRGDRGRG 89
D+ +GRGRGRGRGR RGRG RGRG
Sbjct: 184YADEMIAAGRGRGRGRGRGGGGRGRG-RGRG 273
>TC225776 similar to UP|Q93YH8 (Q93YH8) HMG I/Y like protein, partial (26%)
Length = 1663
Score = 45.1 bits (105), Expect = 4e-05
Identities = 34/100 (34%), Positives = 45/100 (45%), Gaps = 9/100 (9%)
Frame = +1
Query: 9 GGGRGRGKPLEEAAQEAP--------QAPVVNRHIRVRQT-PADAESDNVPRRQPMNRFV 59
G R G+P + +Q P A V ++ +T P DA + PR + +
Sbjct: 523 GPKRRPGRPPKNQSQPTPIPFAPADAAAAVGTEYVAAAETVPVDAAAALGPRARGRPKKY 702
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRG 99
D+ +GRGRGRG RGRGG GGRG GRG
Sbjct: 703 ADEMVAAGRGRGRG------------RGRGGGGGRGRGRG 786
>TC225230 similar to UP|Q9SZZ1 (Q9SZZ1) Fibrillarin-like protein (Fibrillarin
2) (AtFib2), partial (66%)
Length = 773
Score = 44.7 bits (104), Expect = 5e-05
Identities = 34/81 (41%), Positives = 40/81 (48%), Gaps = 2/81 (2%)
Frame = +1
Query: 61 DDGDG-SGRGRGRGRGRDVYARGRGDRGRGGRGG-RGDGRGGFKRYGDDRTSHQDIARSN 118
D G G G GRG RG ARG G RG GGRGG RG GRGG R G S +
Sbjct: 91 DRGRGRGGGGRGGDRGTPFKARG-GGRGGGGRGGGRGGGRGG-GRGGMKGGSKVVVQPHR 264
Query: 119 ADGLYVGDNADGEKLAKKLGP 139
DG+++ + + K L P
Sbjct: 265 HDGIFIAKGKEDALVTKNLVP 327
>TC232422 weakly similar to UP|Q41645 (Q41645) Extensin (Fragment), partial
(11%)
Length = 637
Score = 43.5 bits (101), Expect = 1e-04
Identities = 29/77 (37%), Positives = 35/77 (44%), Gaps = 12/77 (15%)
Frame = -3
Query: 36 RVRQTPADAESDNVPRRQP--------MNRFVRDDGDGSG----RGRGRGRGRDVYARGR 83
R+ Q A + P RQP + R ++G G G R RG G G RG
Sbjct: 269 RIEQKRALRATRGHPMRQPRLLRLWGCLERVKVEEGQGGGEEDERDRGGGGGEGRIGRGG 90
Query: 84 GDRGRGGRGGRGDGRGG 100
G GR GRGG G+GR G
Sbjct: 89 GGEGRTGRGGGGEGRTG 39
Score = 35.8 bits (81), Expect = 0.025
Identities = 22/42 (52%), Positives = 22/42 (52%), Gaps = 1/42 (2%)
Frame = -3
Query: 60 RDDGDGSGRGR-GRGRGRDVYARGRGDRGRGGRGGRGDGRGG 100
RD G G G GR GRG G G G GRGG G GRGG
Sbjct: 137 RDRGGGGGEGRIGRGGG------GEGRTGRGGGGEGRTGRGG 30
>TC221563 similar to UP|O23702 (O23702) Dehydrogenase, partial (21%)
Length = 486
Score = 43.5 bits (101), Expect = 1e-04
Identities = 26/51 (50%), Positives = 26/51 (50%)
Frame = -3
Query: 50 PRRQPMNRFVRDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGG 100
PRR R R G G RGR R RGR RG G R GG G RG GR G
Sbjct: 382 PRRARCRRG-RGGGRGQRRGRSRRRGRRTQGRG*GGRSGGGAGRRGGGRRG 233
>TC217547 weakly similar to UP|Q61869 (Q61869) Keratin 2 epidermis, partial
(8%)
Length = 700
Score = 42.0 bits (97), Expect = 4e-04
Identities = 28/67 (41%), Positives = 33/67 (48%)
Frame = +1
Query: 65 GSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDRTSHQDIARSNADGLYV 124
G GRGRGRGRGR Y RGD R + G GG R+G + T D SN G V
Sbjct: 106 GGGRGRGRGRGRGSY-NNRGDNSNNER--QDSGYGG--RWGSNNTKDSDDGLSNFPGAKV 270
Query: 125 GDNADGE 131
++ E
Sbjct: 271 QNSPGRE 291
>TC224592 similar to UP|Q93VA8 (Q93VA8) At1g76010/T4O12_22, partial (63%)
Length = 1120
Score = 41.6 bits (96), Expect = 5e-04
Identities = 28/68 (41%), Positives = 30/68 (43%), Gaps = 1/68 (1%)
Frame = +1
Query: 39 QTPADAESDNVPRRQPMNRFVRDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGR 98
Q A + D P R G G GRGRG G Y G DR RG GRG GR
Sbjct: 484 QVKAATDFDYEGEGSPNGRVRGRGGRGRGRGRGNGFISADYEDGGWDRNRGNARGRGRGR 663
Query: 99 G-GFKRYG 105
G GF+ G
Sbjct: 664 GRGFRGRG 687
Score = 41.6 bits (96), Expect = 5e-04
Identities = 26/59 (44%), Positives = 29/59 (49%), Gaps = 14/59 (23%)
Frame = +1
Query: 61 DDGDGSGRGRGRGRGRDVYARGRG-------DRGRGGRG-------GRGDGRGGFKRYG 105
D G+ RGRGRGRGR RGRG D GG GRG GRGG++ G
Sbjct: 622 DRNRGNARGRGRGRGRGFRGRGRGGYNGPHVDMQDGGYNQDVPQGRGRGRGRGGYRGRG 798
Score = 29.3 bits (64), Expect = 2.4
Identities = 23/60 (38%), Positives = 25/60 (41%)
Frame = +1
Query: 62 DGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDRTSHQDIARSNADG 121
D D G G GR RGRG RGRG G G ++ G DR R NA G
Sbjct: 502 DFDYEGEGSPNGR-----VRGRGGRGRGRGRGNGFISADYEDGGWDRN------RGNARG 648
Score = 27.7 bits (60), Expect = 6.9
Identities = 25/81 (30%), Positives = 33/81 (39%)
Frame = +1
Query: 9 GGGRGRGKPLEEAAQEAPQAPVVNRHIRVRQTPADAESDNVPRRQPMNRFVRDDGDGSGR 68
G GRGRG+ + P H+ ++ + +VP+ G G GR
Sbjct: 646 GRGRGRGRGFRGRGRGGYNGP----HVDMQD---GGYNQDVPQ-----------GRGRGR 771
Query: 69 GRGRGRGRDVYARGRGDRGRG 89
GRG RG RGRG R G
Sbjct: 772 GRGGYRG-----RGRGFRSNG 819
>BU765146 similar to GP|21322711|e pherophorin-dz1 protein {Volvox carteri f.
nagariensis}, partial (25%)
Length = 407
Score = 40.8 bits (94), Expect = 8e-04
Identities = 20/38 (52%), Positives = 21/38 (54%)
Frame = +2
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGG 100
G G G G G G G + RG GRGGRGG G G GG
Sbjct: 167 GGGGGGGGGGGGGGGGFFRGERXXGRGGRGGGGGGGGG 280
Score = 37.4 bits (85), Expect = 0.009
Identities = 19/38 (50%), Positives = 20/38 (52%)
Frame = +2
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGG 100
G G G GRG G G G G G+GG GG G G GG
Sbjct: 80 GGGGGGGRGGGGGGGGGGGGGGGGGKGGGGGGGGGGGG 193
Score = 35.4 bits (80), Expect = 0.033
Identities = 18/36 (50%), Positives = 19/36 (52%)
Frame = +1
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGR 98
G G G G G G G RG G G GG GG G+GR
Sbjct: 274 GGGGGGGGGXGGGGGGGGRGGGGGGGGGGGGGGEGR 381
Score = 35.0 bits (79), Expect = 0.043
Identities = 20/45 (44%), Positives = 22/45 (48%)
Frame = +3
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDD 107
G G G G GRG G G G GG GG G G GG +R G +
Sbjct: 288 GGGGXGGGGGGRG-----EGGGGGGGGGGGGGGGGEGGGRRGGGE 407
Score = 34.7 bits (78), Expect = 0.057
Identities = 18/37 (48%), Positives = 18/37 (48%)
Frame = +1
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRG 99
G G G G G G G GRG G GG GG G G G
Sbjct: 268 GGGGGGGGGGGXGGGGGGGGRGGGGGGGGGGGGGGEG 378
Score = 34.7 bits (78), Expect = 0.057
Identities = 20/39 (51%), Positives = 20/39 (51%), Gaps = 1/39 (2%)
Frame = +2
Query: 63 GDGSGRGRGRGRGRDVYARG-RGDRGRGGRGGRGDGRGG 100
G G G G G RG RG RG G GG GG G G GG
Sbjct: 188 GGGGGGGGGFFRGERXXGRGGRGGGGGGGGGGGGGGGGG 304
Score = 32.3 bits (72), Expect = 0.28
Identities = 19/38 (50%), Positives = 21/38 (55%)
Frame = +3
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGG 100
G+G G G G G G G G G GG GGRG+G GG
Sbjct: 249 GEGGGGGGGGGGG------GGGGXGGGG-GGRGEGGGG 341
Score = 31.2 bits (69), Expect = 0.63
Identities = 18/39 (46%), Positives = 18/39 (46%)
Frame = +3
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGF 101
G G G GRG G G G GG GG G G GGF
Sbjct: 135 GGGGGGGRGGGGG-----------GGGGGGGGGGGGGGF 218
>TC208462 homologue to UP|LSM4_TOBAC (Q43582) Probable U6 snRNA-associated
Sm-like protein LSm4 (Glycine-rich protein 10) (GRP 10),
partial (94%)
Length = 794
Score = 40.4 bits (93), Expect = 0.001
Identities = 23/46 (50%), Positives = 28/46 (60%), Gaps = 6/46 (13%)
Frame = +3
Query: 60 RDDGDGSGRGRGRGRGRDVYA-RGRGDRGRGGRGG-----RGDGRG 99
R+DG G + +G GRG D +G+G RGRGG GG RG GRG
Sbjct: 432 REDGPGGRQPKGIGRGLDEGGPKGQGGRGRGGPGGKPGGNRGGGRG 569
Score = 36.6 bits (83), Expect = 0.015
Identities = 25/64 (39%), Positives = 30/64 (46%), Gaps = 18/64 (28%)
Frame = +3
Query: 60 RDDGDGSGRGRGRGRGR-----------------DVYARGRGDRGRGGRGGR-GDGRGGF 101
R D G GRGRGRGR + +G+G RGRGG GG+ G RGG
Sbjct: 384 RTDRKPPGVGRGRGRGREDGPGGRQPKGIGRGLDEGGPKGQGGRGRGGPGGKPGGNRGGG 563
Query: 102 KRYG 105
+ G
Sbjct: 564 RGRG 575
>TC235092 similar to UP|O91332 (O91332) EBNA-1, partial (5%)
Length = 641
Score = 40.4 bits (93), Expect = 0.001
Identities = 25/43 (58%), Positives = 28/43 (64%), Gaps = 2/43 (4%)
Frame = -2
Query: 70 RGRGRGRDVYARGRGDRGR--GGRGGRGDGRGGFKRYGDDRTS 110
RGR GR + AR RG RGR G RGGRG+GR G +R G R S
Sbjct: 187 RGRVSGRILGAR-RGARGRSRGRRGGRGEGRSGRRRNGQGRRS 62
Score = 28.1 bits (61), Expect = 5.3
Identities = 17/50 (34%), Positives = 23/50 (46%)
Frame = -2
Query: 65 GSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDRTSHQDI 114
G RGR GRG GR R R G+G R G+ R + + +D+
Sbjct: 139 GRSRGRRGGRG-----EGRSGRRRNGQGRRSSGKWRVFRVLERLVAQEDL 5
>TC234067 weakly similar to UP|Q9SBM1 (Q9SBM1) Hydroxyproline-rich
glycoprotein DZ-HRGP precursor, partial (8%)
Length = 614
Score = 39.3 bits (90), Expect = 0.002
Identities = 31/80 (38%), Positives = 39/80 (48%), Gaps = 4/80 (5%)
Frame = -1
Query: 28 APVVNRHIRVRQTPA---DAESDNVPRRQPMNRFVRDDGDGSGRGRGRGRGRDVYARGRG 84
+P++NR V + + DAE+ + F GDG R R RG G A GRG
Sbjct: 299 SPLINRACSVSSSSSSQNDAETTDERFDHKTFTFEHHGGDGGERERLRGGG----A*GRG 132
Query: 85 D-RGRGGRGGRGDGRGGFKR 103
D GRGG G G+ RGG R
Sbjct: 131 DGAGRGGGEGGGERRGGSAR 72
>TC210426 similar to UP|Q9FK53 (Q9FK53) Similarity to GAR1 protein, partial
(50%)
Length = 434
Score = 38.9 bits (89), Expect = 0.003
Identities = 25/56 (44%), Positives = 26/56 (45%)
Frame = +3
Query: 53 QPMNRFVRDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDR 108
QP + G G GRG GRG GRG G GRGG GRGG R G R
Sbjct: 210 QPKGQSAGRGGGGGGRGGGRG--------GRGGGGFRGRGGPRGGRGGPPRGGGFR 353
Score = 37.0 bits (84), Expect = 0.011
Identities = 24/48 (50%), Positives = 28/48 (58%), Gaps = 1/48 (2%)
Frame = +3
Query: 54 PMNRFV-RDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGG 100
P+ RF+ + G +GRG G G GR GRG RG GG GRG RGG
Sbjct: 189 PLARFLPQPKGQSAGRGGGGG-GR---GGGRGGRGGGGFRGRGGPRGG 320
>TC230571 similar to UP|Q41645 (Q41645) Extensin (Fragment), partial (7%)
Length = 667
Score = 38.9 bits (89), Expect = 0.003
Identities = 25/40 (62%), Positives = 26/40 (64%), Gaps = 1/40 (2%)
Frame = -2
Query: 67 GRGRGRGRGRDVYARGRGD-RGRGGRGGRGDGRGGFKRYG 105
GRGR RGRG RGRG RGRGG GRG GG+KR G
Sbjct: 357 GRGRRRGRG----GRGRGKARGRGGCPGRG---GGWKRCG 259
Score = 29.6 bits (65), Expect = 1.8
Identities = 17/30 (56%), Positives = 20/30 (66%), Gaps = 3/30 (10%)
Frame = -2
Query: 67 GRGRGRGRGR-DVYARGRGDR--GRGGRGG 93
GRGRG+ RGR RG G + G+GGRGG
Sbjct: 330 GRGRGKARGRGGCPGRGGGWKRCGQGGRGG 241
>TC225232 similar to UP|Q9SZZ1 (Q9SZZ1) Fibrillarin-like protein (Fibrillarin
2) (AtFib2), partial (83%)
Length = 1065
Score = 38.9 bits (89), Expect = 0.003
Identities = 28/80 (35%), Positives = 35/80 (43%)
Frame = +3
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDRTSHQDIARSNA 119
R DGD + RG GR G RG G GGRG GRGG K G + Q
Sbjct: 24 RKDGDRGTPFKARGGGR-------GGRGGGRGGGRGGGRGGMK--GGSKVVVQP---HRH 167
Query: 120 DGLYVGDNADGEKLAKKLGP 139
DG+++ + + K L P
Sbjct: 168 DGIFIAKGKEDALVTKNLVP 227
>BM086617 homologue to GP|9651941|gb receptor-like protein kinase 1 {Glycine
max}, partial (12%)
Length = 428
Score = 38.1 bits (87), Expect = 0.005
Identities = 24/51 (47%), Positives = 28/51 (54%), Gaps = 4/51 (7%)
Frame = -3
Query: 63 GDGSGRGRGRGRG---RDVYARGRGDRGR-GGRGGRGDGRGGFKRYGDDRT 109
G G RGR R G R Y RGRG RGR GGRGG + R ++ G R+
Sbjct: 408 GSGGRRGRRRKPGWGRRTGYRRGRGGRGRGGGRGGHREWRREIRKLG*GRS 256
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.137 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,646,825
Number of Sequences: 63676
Number of extensions: 128003
Number of successful extensions: 2874
Number of sequences better than 10.0: 375
Number of HSP's better than 10.0 without gapping: 1698
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2327
length of query: 310
length of database: 12,639,632
effective HSP length: 97
effective length of query: 213
effective length of database: 6,463,060
effective search space: 1376631780
effective search space used: 1376631780
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC147483.4


