

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147483.2 + phase: 0
(384 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
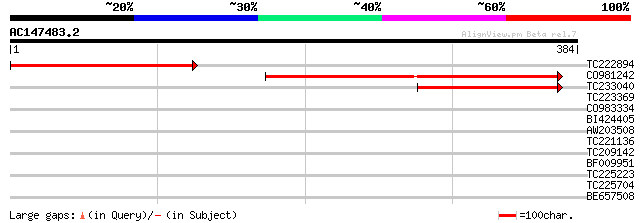
Score E
Sequences producing significant alignments: (bits) Value
TC222894 221 6e-58
CO981242 179 1e-45
TC233040 123 1e-28
TC223369 40 0.001
CO983334 37 0.020
BI424405 35 0.074
AW203508 weakly similar to SP|P57758|CTNS_ Cystinosin homolog. [... 31 0.82
TC221136 homologue to UP|LBD4_ARATH (Q9SHE9) LOB domain protein ... 30 2.4
TC209142 similar to UP|Q8YY49 (Q8YY49) All1003 protein, partial ... 29 3.1
BF009951 28 5.3
TC225223 similar to UP|Q6QLN2 (Q6QLN2) Endo-1,4-beta-glucanase ... 28 5.3
TC225704 homologue to UP|Q9ZRD8 (Q9ZRD8) GMFP5 (Fragment), parti... 28 5.3
BE657508 homologue to SP|Q42783|BCCP_ Biotin carboxyl carrier pr... 28 9.1
>TC222894
Length = 537
Score = 221 bits (562), Expect = 6e-58
Identities = 102/127 (80%), Positives = 110/127 (86%)
Frame = +3
Query: 1 MAPSYCVKENKECVKWVETYFKDCLCNLRDDISFSLGLMSLVSWGVAEIPQIITIFRNKS 60
M PSYCVKE K CV+WVE YFKDCLCNL+D+ISFS GL SLV WGVAEIPQIITIFR K
Sbjct: 150 MPPSYCVKERKPCVRWVEKYFKDCLCNLKDEISFSFGLTSLVFWGVAEIPQIITIFRTKK 329
Query: 61 SHGISLAFLLTWVAGDICNLVGCLLEPATLPTQFYTALLYASTTIILLLQIVYYDHILRW 120
SHG+SL FLLTWVAGDICNL GC+LEPATLPTQ+YTALLY TTI+LLL IVYYD+I RW
Sbjct: 330 SHGVSLVFLLTWVAGDICNLTGCILEPATLPTQYYTALLYTITTIVLLLLIVYYDYISRW 509
Query: 121 CKHRQNV 127
KHRQ V
Sbjct: 510 YKHRQKV 530
>CO981242
Length = 847
Score = 179 bits (455), Expect = 1e-45
Identities = 106/201 (52%), Positives = 133/201 (65%)
Frame = -1
Query: 174 GVLLHVSKIVSGECNSTLVYPSKSC*KWSFSLGIHPRFF****SISSYVQYLYNQALVHT 233
G LL+VSKI+ E +STL SC*KW S GI F *S S +++ N + +
Sbjct: 844 GALLYVSKIIGWEWHSTLGNLHGSC*KWPCSHGIDQ*LFFRQ*STSCFLK*FCNPSYANP 665
Query: 234 TFCRWTLWNISSYSH*LALKG*FNEIWIHRIYWNKTP*GICCGT*YIWTIFGMDHGCYIH 293
TFC W LWNISS * A +G E IHRI+W KT G+ T*YIW++ GM HGCY++
Sbjct: 664 TFCCWKLWNISSSCC*FASQGQCFEGRIHRIWWKKTFAGVR-DT*YIWSMAGMAHGCYLY 488
Query: 294 M*SNSSNMVEH*KR*CRGLKSFHVRFCTDC*YFLRGKYSCTNN*V*KHQSKFAMVVGCHS 353
S SSNMVEH*KR C G+KSFHVR C* LRGKYSC N + K+QS +AMV+GC +
Sbjct: 487 KWSCSSNMVEH*KRQCGGVKSFHVRLRIGC*CHLRGKYSCKNYRMGKNQS*YAMVIGCRN 308
Query: 354 LRGTRLFYNIAVHLL*ILSQQ 374
LRG L +NI+V+LL*+LS++
Sbjct: 307 LRGIGLLHNISVYLL*MLSKE 245
>TC233040
Length = 725
Score = 123 bits (309), Expect = 1e-28
Identities = 61/98 (62%), Positives = 77/98 (78%)
Frame = +1
Query: 277 T*YIWTIFGMDHGCYIHM*SNSSNMVEH*KR*CRGLKSFHVRFCTDC*YFLRGKYSCTNN 336
T*YIW++ GM HGCY++ S+SSNMVE+*K+ C GLKSFHVR C* LRGKYSC N+
Sbjct: 13 T*YIWSMAGMAHGCYLYKWSSSSNMVEY*KKHCGGLKSFHVRLRIGC*CHLRGKYSCKNH 192
Query: 337 *V*KHQSKFAMVVGCHSLRGTRLFYNIAVHLL*ILSQQ 374
+ KHQS +AMV+GC +LRG F+NIA+HLL ILS++
Sbjct: 193 RMGKHQS*YAMVIGCRNLRGIGHFHNIAIHLLQILSKE 306
>TC223369
Length = 706
Score = 40.4 bits (93), Expect = 0.001
Identities = 26/81 (32%), Positives = 43/81 (52%)
Frame = +2
Query: 291 YIHM*SNSSNMVEH*KR*CRGLKSFHVRFCTDC*YFLRGKYSCTNN*V*KHQSKFAMVVG 350
YI +SN+ ++ KR C G +SF V C + L +Y+C + K+Q++FAM
Sbjct: 125 YIFGWKTASNLFKYQKRSC*GTQSFDVPVCCNWECHLCSEYTCDQLGLVKNQTEFAMASR 304
Query: 351 CHSLRGTRLFYNIAVHLL*IL 371
C + + FY AV++ +L
Sbjct: 305 CWRVCPS*FFYFNAVYIFSLL 367
>CO983334
Length = 807
Score = 36.6 bits (83), Expect = 0.020
Identities = 21/61 (34%), Positives = 33/61 (53%)
Frame = -2
Query: 311 GLKSFHVRFCTDC*YFLRGKYSCTNN*V*KHQSKFAMVVGCHSLRGTRLFYNIAVHLL*I 370
G +SF V C + L +YSC + K+Q++FAM C + +R FY AV++ +
Sbjct: 617 GTQSFDVPVCCNWECHLCSEYSCDQPGLVKNQTEFAMASRCWGMCPSRFFYFNAVYIFSL 438
Query: 371 L 371
L
Sbjct: 437 L 435
>BI424405
Length = 421
Score = 34.7 bits (78), Expect = 0.074
Identities = 16/33 (48%), Positives = 22/33 (66%)
Frame = +1
Query: 36 LGLMSLVSWGVAEIPQIITIFRNKSSHGISLAF 68
LG ++ +SW VA PQ+I FR KS G+SL +
Sbjct: 112 LGWLAFLSWSVAGYPQLILNFRRKSVVGLSLDY 210
>AW203508 weakly similar to SP|P57758|CTNS_ Cystinosin homolog. [Mouse-ear
cress] {Arabidopsis thaliana}, partial (35%)
Length = 348
Score = 31.2 bits (69), Expect = 0.82
Identities = 26/65 (40%), Positives = 33/65 (50%), Gaps = 3/65 (4%)
Frame = +2
Query: 36 LGLMSLVSWGVAEIPQIITIFRNKSSHGISLAFLLTWVAGDICNLV---GCLLEPATLPT 92
LG ++ +SW VA PQ+I R KS G SL + +I NL L+ ATLP
Sbjct: 77 LGWLAYLSWSVAGYPQLILNCRRKSVVGSSLDY-------EILNLTKHFSYLIYNATLP- 232
Query: 93 QFYTA 97
F TA
Sbjct: 233 -FVTA 244
>TC221136 homologue to UP|LBD4_ARATH (Q9SHE9) LOB domain protein 4, partial
(44%)
Length = 607
Score = 29.6 bits (65), Expect = 2.4
Identities = 14/33 (42%), Positives = 20/33 (60%)
Frame = -2
Query: 60 SSHGISLAFLLTWVAGDICNLVGCLLEPATLPT 92
+S+ I L L W +G+ CN++ LL P TL T
Sbjct: 111 ASYTIELTASLLWCSGNSCNILFTLLAPKTL*T 13
>TC209142 similar to UP|Q8YY49 (Q8YY49) All1003 protein, partial (15%)
Length = 918
Score = 29.3 bits (64), Expect = 3.1
Identities = 24/92 (26%), Positives = 44/92 (47%), Gaps = 5/92 (5%)
Frame = -3
Query: 72 WVAGDICNLVGCLLEP---ATLPTQFYTALLYASTTIILL--LQIVYYDHILRWCKHRQN 126
+VAG N G P P +F A + A + ++ + L++++ +L WC +
Sbjct: 328 FVAGHRANAGGFFCTPR*SGFFPPKFKMAPMVAISLLLNMSSLRLIWSVGVLSWCF*QNT 149
Query: 127 VKSKLLLLQLYYLTG**RREKTLESKTKSSVL 158
++ + LLL L L R +K LE ++ V+
Sbjct: 148 IRGRELLLPLLGLVMNLRLQKELEDAIEAIVV 53
>BF009951
Length = 283
Score = 28.5 bits (62), Expect = 5.3
Identities = 15/37 (40%), Positives = 19/37 (50%)
Frame = -1
Query: 176 LLHVSKIVSGECNSTLVYPSKSC*KWSFSLGIHPRFF 212
L VS +S L+YP + C* W +SLG FF
Sbjct: 274 LRRVSSPYLSNRSSRLLYPPRLC*GWKWSLGWR*EFF 164
>TC225223 similar to UP|Q6QLN2 (Q6QLN2) Endo-1,4-beta-glucanase , partial
(61%)
Length = 1470
Score = 28.5 bits (62), Expect = 5.3
Identities = 12/43 (27%), Positives = 19/43 (43%)
Frame = -2
Query: 159 WDCNTKWHTKRSSSRGVLLHVSKIVSGECNSTLVYPSKSC*KW 201
W C W R +GV ++ ++G+C+ SC* W
Sbjct: 1112 WWCGKHWGNSRE--KGVFVYTGGFITGQCHKCSY*TCISC*SW 990
>TC225704 homologue to UP|Q9ZRD8 (Q9ZRD8) GMFP5 (Fragment), partial (66%)
Length = 821
Score = 28.5 bits (62), Expect = 5.3
Identities = 12/39 (30%), Positives = 20/39 (50%)
Frame = +1
Query: 144 RREKTLESKTKSSVLWDCNTKWHTKRSSSRGVLLHVSKI 182
R+ KTL + + W+ T+WH +RS + V +I
Sbjct: 151 RKRKTLLQRRRLLTKWNICTRWHLRRSGTTEDTFQVKQI 267
>BE657508 homologue to SP|Q42783|BCCP_ Biotin carboxyl carrier protein of
acetyl-CoA carboxylase chloroplast precursor (BCCP).
[Soybean], partial (11%)
Length = 711
Score = 27.7 bits (60), Expect = 9.1
Identities = 13/57 (22%), Positives = 29/57 (50%)
Frame = -2
Query: 82 GCLLEPATLPTQFYTALLYASTTIILLLQIVYYDHILRWCKHRQNVKSKLLLLQLYY 138
GC++ ++ F+ LYA + I+++ +V C + +KS+ + +LY+
Sbjct: 527 GCIMVDISIQQTFH*TKLYAISLILMICLLV--------CNYEMQIKSRKEMFRLYH 381
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.354 0.155 0.589
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,083,363
Number of Sequences: 63676
Number of extensions: 400213
Number of successful extensions: 4533
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 4475
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4529
length of query: 384
length of database: 12,639,632
effective HSP length: 99
effective length of query: 285
effective length of database: 6,335,708
effective search space: 1805676780
effective search space used: 1805676780
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC147483.2


