

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147482.3 - phase: 0
(175 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
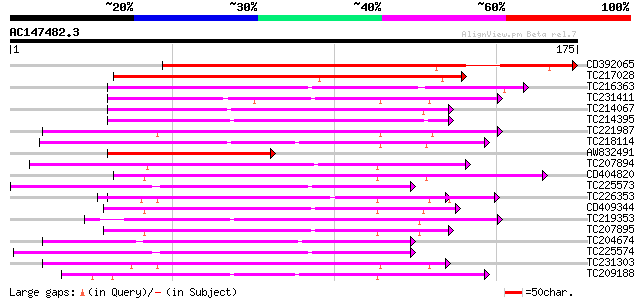
Score E
Sequences producing significant alignments: (bits) Value
CD392065 163 3e-41
TC217028 weakly similar to UP|Q39370 (Q39370) Lamin, partial (80%) 115 1e-26
TC216363 similar to UP|Q7YHL4 (Q7YHL4) NADH dehydrogenase subuni... 107 3e-24
TC231411 similar to GB|AAP21370.1|30102904|BT006562 At3g27200 {A... 70 4e-13
TC214067 weakly similar to UP|Q949E8 (Q949E8) Uclacyanin 3-like ... 70 5e-13
TC214395 weakly similar to UP|Q949E8 (Q949E8) Uclacyanin 3-like ... 69 1e-12
TC221987 nodulin 69 1e-12
TC218114 similar to UP|BCP_PEA (Q41001) Blue copper protein prec... 67 3e-12
AW832491 65 2e-11
TC207894 similar to UP|O81500 (O81500) F9D12.16 protein (At5g263... 63 6e-11
CD404820 weakly similar to SP|Q41001|BCP_P Blue copper protein p... 63 8e-11
TC225573 similar to UP|Q9ZRV5 (Q9ZRV5) Basic blue copper protein... 62 1e-10
TC226353 weakly similar to UP|Q9M510 (Q9M510) Dicyanin, partial ... 62 1e-10
CD409344 62 1e-10
TC219353 weakly similar to UP|Q8L555 (Q8L555) Uclacyanin 3-like ... 62 1e-10
TC207895 similar to UP|O81500 (O81500) F9D12.16 protein (At5g263... 62 1e-10
TC204674 similar to UP|Q9ZRV5 (Q9ZRV5) Basic blue copper protein... 61 2e-10
TC225574 similar to UP|Q9ZRV5 (Q9ZRV5) Basic blue copper protein... 61 3e-10
TC231303 homologue to UP|N552_SOYBN (Q02917) Early nodulin 55-2 ... 57 4e-09
TC209188 similar to UP|BCP_PEA (Q41001) Blue copper protein prec... 57 6e-09
>CD392065
Length = 602
Score = 163 bits (413), Expect = 3e-41
Identities = 82/136 (60%), Positives = 103/136 (75%), Gaps = 8/136 (5%)
Frame = -2
Query: 48 WSSQEHVYVGDWLKFVFDKRYYNVLEVNKTGYDYCIDMTFIRNLTRGGRDVVQLTEAKTY 107
W++QEHVYVG WL F FD+RY+NVLEVNKT Y+ CID FI+N+TRGGRDVVQ+TEA+TY
Sbjct: 598 WAAQEHVYVGYWLIFKFDRRYFNVLEVNKTSYENCIDRDFIKNITRGGRDVVQMTEARTY 419
Query: 108 YFITGGGYCFHGMKVAVDVQEHP-------TPAPSPSLSDTAKSGGDSILPSMYTCFGII 160
Y+++ GGYCFHGMKVAV VQE+ PAPSP +S + S++TC II
Sbjct: 418 YYLSDGGYCFHGMKVAVQVQEYQDPALAMVAPAPSPVVSGS----------SVFTCIWII 269
Query: 161 VANVV-YVSLVLVGIL 175
VANVV +V+L++VGIL
Sbjct: 268 VANVVLFVNLMVVGIL 221
>TC217028 weakly similar to UP|Q39370 (Q39370) Lamin, partial (80%)
Length = 862
Score = 115 bits (287), Expect = 1e-26
Identities = 57/113 (50%), Positives = 74/113 (65%), Gaps = 4/113 (3%)
Frame = +1
Query: 33 VGGSKGWKENVNYTTWSSQEHVYVGDWLKFVFDKRYYNVLEVNKTGYDYCIDMTFIRNLT 92
VG ++ W N+NYT W+ +H Y+GDWL FV+D+ +VLEVNKT Y+ C + N T
Sbjct: 181 VGNNQFWNPNINYTEWAKGKHFYLGDWLYFVYDRNQASVLEVNKTDYETCNSDHPLTNWT 360
Query: 93 RG-GRDVVQLTEAKTYYFITGGGYCFHGMKVAVDVQEHPTP---APSPSLSDT 141
RG GRDVV L KTYY I+G G+CF GMK+AV V++ P P AP S + T
Sbjct: 361 RGAGRDVVPLNVTKTYYIISGRGFCFSGMKIAVHVEKLPPPPKAAPVKSAAPT 519
>TC216363 similar to UP|Q7YHL4 (Q7YHL4) NADH dehydrogenase subunit 1, partial
(5%)
Length = 854
Score = 107 bits (267), Expect = 3e-24
Identities = 58/143 (40%), Positives = 74/143 (51%), Gaps = 13/143 (9%)
Frame = +1
Query: 31 HKVGGSKGWKENVNYTTWSSQEHVYVGDWLKFVFDKRYYNVLEVNKTGYDYCIDMTFIRN 90
H VG ++GW NYT W++ YVGD + F + K YNV EVN+TGYD C + N
Sbjct: 196 HIVGANRGWNPGFNYTLWANNHTFYVGDLISFRYQKNQYNVFEVNQTGYDNCTTEGAVGN 375
Query: 91 LTRGGRDVVQLTEAKTYYFITGGGYCFHGMKVAVDVQEHPTPAPSPSLSDTAKSGGDSIL 150
+ G+D + L +AK YYFI G G CF GMKV+V V HP P P S S S
Sbjct: 376 WS-SGKDFIPLNKAKRYYFICGNGQCFSGMKVSVVV--HPLPPPPTSAVAAQHSSPKSAS 546
Query: 151 P-------------SMYTCFGII 160
P ++ CFGI+
Sbjct: 547 PVLLKRGFRSLLVSTVSACFGIV 615
>TC231411 similar to GB|AAP21370.1|30102904|BT006562 At3g27200 {Arabidopsis
thaliana;} , partial (57%)
Length = 855
Score = 70.5 bits (171), Expect = 4e-13
Identities = 43/126 (34%), Positives = 75/126 (59%), Gaps = 4/126 (3%)
Frame = +2
Query: 31 HKVGGSKGWKENVNYTTWSSQEHVYVGDWLKFVFDKRYYNVLEV-NKTGYDYCIDMTFIR 89
H VGGS+GW ++ ++ +W+S + VGD L F + +++V+E+ N++ Y C + ++
Sbjct: 131 HVVGGSQGWDQSTDFKSWTSGQTFKVGDKLVFKYSS-FHSVVELGNESAYKNCDISSPVQ 307
Query: 90 NLTRGGRDVVQLTEAKTYYFITGG-GYCFHGMKVAVDVQE--HPTPAPSPSLSDTAKSGG 146
+L+ G DVV+L + T YF G G+C GMKV + +++ P+PA SP+ S +
Sbjct: 308 SLST-GNDVVKLDKPGTRYFTCGTLGHCSQGMKVKITIRKGNAPSPALSPATSPSLSPSL 484
Query: 147 DSILPS 152
+L S
Sbjct: 485 SPLLSS 502
>TC214067 weakly similar to UP|Q949E8 (Q949E8) Uclacyanin 3-like protein,
partial (50%)
Length = 948
Score = 70.1 bits (170), Expect = 5e-13
Identities = 38/108 (35%), Positives = 57/108 (52%), Gaps = 1/108 (0%)
Frame = +1
Query: 31 HKVGGSKGWKENVNYTTWSSQEHVYVGDWLKFVFDKRYYNVLEVNKTGYDYCIDMTFIRN 90
H+VG + GW VNY TW+S + VGD L F +D + V EV+++GY+ C I+N
Sbjct: 157 HEVGDTSGWALGVNYNTWASGKTFTVGDTLVFKYDST-HQVDEVDESGYNSCSSSNSIKN 333
Query: 91 LTRGGRDVVQLTEAKTYYFITGGGYCFHGMKVAVDV-QEHPTPAPSPS 137
G + + K Y+ G+C GMK+ ++V TP +PS
Sbjct: 334 YQDGNSKIELTSPGKRYFLCPISGHCAGGMKLQINVAAASGTPPTTPS 477
>TC214395 weakly similar to UP|Q949E8 (Q949E8) Uclacyanin 3-like protein,
partial (44%)
Length = 652
Score = 68.9 bits (167), Expect = 1e-12
Identities = 37/107 (34%), Positives = 57/107 (52%)
Frame = +2
Query: 31 HKVGGSKGWKENVNYTTWSSQEHVYVGDWLKFVFDKRYYNVLEVNKTGYDYCIDMTFIRN 90
H+VG + GW VNY TW+S + +GD L F +D + V EV+++GY+ C I+N
Sbjct: 14 HEVGDTGGWALGVNYNTWASGKTFRIGDNLVFKYDSTH-QVDEVDESGYNSCSSSNIIKN 190
Query: 91 LTRGGRDVVQLTEAKTYYFITGGGYCFHGMKVAVDVQEHPTPAPSPS 137
G + + K Y+ G+C GMK+ ++V TP +PS
Sbjct: 191 YKDGNTKIELTSTGKRYFLCPISGHCAGGMKLQINVVA-GTPPTTPS 328
>TC221987 nodulin
Length = 844
Score = 68.9 bits (167), Expect = 1e-12
Identities = 50/167 (29%), Positives = 80/167 (46%), Gaps = 25/167 (14%)
Frame = +1
Query: 11 SVVLGVLSLWPMVVMGGPKLHKVGGSKGWKENVN----YTTWSSQEHVYVGDWLKFVFDK 66
S L +LS+W ++ + + VGGS+ WK ++ + W+S +GD L F +D+
Sbjct: 40 SPFLFMLSMWLLISISEAAKYVVGGSETWKFPLSKPDSLSHWASSHRFKIGDTLIFKYDE 219
Query: 67 RYYNVLEVNKTGYDYCIDMTFIRNLTRGGRDVVQLTEAKTYYFITGG-GYCFHGMKVAVD 125
R +V EVN+T Y+ C + L G V LT++ +FI+G +C G+K+ V
Sbjct: 220 RTESVHEVNETDYEQCNTVGKEHVLFNDGNTKVMLTKSGFRHFISGNQSHCQMGLKLMVV 399
Query: 126 VQEH--------------------PTPAPSPSLSDTAKSGGDSILPS 152
V + P+P+PSPS S + S S LP+
Sbjct: 400 VMSNNTKKKLIHSPSPSSPSPSPSPSPSPSPSPSPSLSSPSPSPLPN 540
>TC218114 similar to UP|BCP_PEA (Q41001) Blue copper protein precursor,
partial (57%)
Length = 901
Score = 67.4 bits (163), Expect = 3e-12
Identities = 47/144 (32%), Positives = 70/144 (47%), Gaps = 5/144 (3%)
Frame = +3
Query: 10 VSVVLGVLSLWPMVVMGGPKLHKVGGSKGWKENVNYTTWSSQEHVYVGDWLKFVFDKRYY 69
V++VLG+ MV+ H VG + GW +Y+TW+S + VGD L F + +
Sbjct: 63 VALVLGLCLALNMVLPTRAATHTVGDTSGWALGADYSTWASGLKLKVGDSLVFNYGAG-H 239
Query: 70 NVLEVNKTGYDYCIDMTFIRNLTRGGRDVVQLTEAKTYYFITGG-GYCFHGMKVAVDVQ- 127
V EV ++ Y C + + G + L A T+YFI G+C GMK+AV V+
Sbjct: 240 TVDEVKESDYKSCTTGNSL-STDSSGTTTITLKTAGTHYFICASPGHCDGGMKLAVKVKA 416
Query: 128 ---EHPTPAPSPSLSDTAKSGGDS 148
P APSP+ D+ D+
Sbjct: 417 KKASAPATAPSPAAKDSPSDSDDT 488
>AW832491
Length = 285
Score = 65.1 bits (157), Expect = 2e-11
Identities = 26/52 (50%), Positives = 33/52 (63%)
Frame = +2
Query: 31 HKVGGSKGWKENVNYTTWSSQEHVYVGDWLKFVFDKRYYNVLEVNKTGYDYC 82
H VG ++GW NYT W++ YVGD + F + K YNV EVN+TGYD C
Sbjct: 17 HIVGANRGWNPGFNYTLWANNHTFYVGDLISFRYQKNQYNVFEVNQTGYDNC 172
>TC207894 similar to UP|O81500 (O81500) F9D12.16 protein (At5g26330), partial
(56%)
Length = 755
Score = 63.2 bits (152), Expect = 6e-11
Identities = 38/139 (27%), Positives = 72/139 (51%), Gaps = 3/139 (2%)
Frame = +2
Query: 7 LLMVSVVLGVLSLWPMVVMGGPKLHKVGGSKGWKE--NVNYTTWSSQEHVYVGDWLKFVF 64
+ +V V+ + + +V + ++KVG S GW ++Y W++ ++ +GD + F +
Sbjct: 47 MTLVERVVVLFIVMTIVKVSYAAVYKVGDSAGWTTLGTIDYRKWAATKNFQIGDTIIFEY 226
Query: 65 DKRYYNVLEVNKTGYDYCIDMTFIRNLTRGGRDVVQLTEAKTYYFITG-GGYCFHGMKVA 123
+ +++NV+ V Y C + I T G +D + +T ++F G G+C G KV
Sbjct: 227 NAKFHNVMRVTHAMYKTCNASSPIATFTTG-KDSINITNHGHHFFFCGVPGHCQAGQKVD 403
Query: 124 VDVQEHPTPAPSPSLSDTA 142
++V + AP+PS S A
Sbjct: 404 INVLKVSAEAPTPSGSALA 460
>CD404820 weakly similar to SP|Q41001|BCP_P Blue copper protein precursor.
[Garden pea] {Pisum sativum}, partial (22%)
Length = 632
Score = 62.8 bits (151), Expect = 8e-11
Identities = 40/145 (27%), Positives = 65/145 (44%), Gaps = 11/145 (7%)
Frame = -3
Query: 33 VGGSKGWK---ENVNYTTWSSQEHVYVGDWLKFVFDKRYYNVLEVNKTGYDYCIDMTFIR 89
VG GW+ + Y W+S ++ VGD L F+F +NV+EV++ Y+ C I
Sbjct: 609 VGDGTGWRVPQDASTYHNWASDKNFTVGDTLSFIFQTGLHNVIEVSEESYNSCSSANPIG 430
Query: 90 NLTRGGRDVVQLTEAKTYYFITG-GGYCFHGMKVAVDVQ-------EHPTPAPSPSLSDT 141
G V L +Y+I G +C +G ++A+ V T AP P S
Sbjct: 429 TTYNTGPANVTLNRGGEHYYICSFGNHCNNGQRLAISVSGSSTAFPPATTTAPPPPSSSA 250
Query: 142 AKSGGDSILPSMYTCFGIIVANVVY 166
S ++LP ++ + +A +Y
Sbjct: 249 PSSFHTTVLPFLFLSAFVFIAF*IY 175
>TC225573 similar to UP|Q9ZRV5 (Q9ZRV5) Basic blue copper protein, partial
(82%)
Length = 863
Score = 62.4 bits (150), Expect = 1e-10
Identities = 36/125 (28%), Positives = 62/125 (48%)
Frame = +1
Query: 1 MKNSRVLLMVSVVLGVLSLWPMVVMGGPKLHKVGGSKGWKENVNYTTWSSQEHVYVGDWL 60
M R + +V+ V+SL ++ + VGG GW N N W + + GD L
Sbjct: 76 MSQGRGSASLPIVVTVVSLLCLLERANAATYSVGGPGGWTFNTN--AWPNGKRFRAGDIL 249
Query: 61 KFVFDKRYYNVLEVNKTGYDYCIDMTFIRNLTRGGRDVVQLTEAKTYYFITGGGYCFHGM 120
F +D +NV+ V+++GY+ C + + G+D ++L + Y+ G+C GM
Sbjct: 250 IFNYDSTTHNVVAVDRSGYNSCKTPGGAKVFS-SGKDQIKLARGQNYFICNYPGHCESGM 426
Query: 121 KVAVD 125
KVA++
Sbjct: 427 KVAIN 441
>TC226353 weakly similar to UP|Q9M510 (Q9M510) Dicyanin, partial (26%)
Length = 1341
Score = 62.4 bits (150), Expect = 1e-10
Identities = 37/114 (32%), Positives = 53/114 (46%), Gaps = 8/114 (7%)
Frame = +3
Query: 31 HKVGGSKGW----KENVNYTTWSSQEHVYVGDWLKFVFDKRYYNVLEVNKTGYDYCIDMT 86
H VG + GW YT W+S + V D L F F +NV +V K+ +D C +
Sbjct: 150 HMVGDATGWIIPAGGAATYTAWASNKTFTVNDTLVFNFATGQHNVAKVTKSAFDACNGGS 329
Query: 87 FIRNLTRGGRDVVQLTEAKTYYFITGGGYCFHGMKVAVDVQE----HPTPAPSP 136
+ LT G V + YY + G +C G K+A++V P+PAP P
Sbjct: 330 AVFTLTSGPATVTLNETGEQYYICSVGSHCSAGQKLAINVNRASSTGPSPAPQP 491
Score = 55.5 bits (132), Expect = 1e-08
Identities = 40/132 (30%), Positives = 62/132 (46%), Gaps = 8/132 (6%)
Frame = +3
Query: 28 PKLHKVGGSKGWKENVN---YTTWSSQEHVYVGDWLKFVFDKRYYNVLEVNKTGYDYCID 84
P VG + GW N YT W+S ++ VGD L F + +NV EV K +D C
Sbjct: 609 PATFIVGETAGWIVPGNASFYTAWASGKNFRVGDVLVFNYASNTHNVEEVTKANFDACSS 788
Query: 85 MTFIRNLTRGGRDVVQLTEAKTYYFITG-GGYCFHGMKVAVDVQEHPTPAP----SPSLS 139
+ I T V L ++ ++FI G G+C G K+A++V T P +P +
Sbjct: 789 ASPIATFTTPPARVT-LNKSGQHFFICGIPGHCLGGQKLAINVTGSSTATPPSAAAPPTT 965
Query: 140 DTAKSGGDSILP 151
++ S ++ P
Sbjct: 966 PSSPSPAGAVTP 1001
>CD409344
Length = 438
Score = 62.4 bits (150), Expect = 1e-10
Identities = 35/118 (29%), Positives = 65/118 (54%), Gaps = 8/118 (6%)
Frame = -2
Query: 30 LHKVGGSKGWK--ENVNYTTWSSQEHVYVGDWLKFVFDKRYYNVLEVNKTGYDYCIDMTF 87
+HKVG S GW N++Y W++ ++ VGD + F ++ +++NV+ V Y C +
Sbjct: 362 VHKVGDSAGWTIIGNIDYKKWAATKNFQVGDTIIFEYNAKFHNVMRVTHGMYKSCNASSP 183
Query: 88 IRNLTRGGRDVVQLTEAKTYYFITG-GGYCFHGMKVAVDV-----QEHPTPAPSPSLS 139
+ ++ G D +++T + F+ G G+C G KV ++V E PTP+P +++
Sbjct: 182 LTRMST-GNDTIKITNYGHHLFLCGVPGHCQAGQKVDINVVKKVSAEAPTPSPISAMA 12
>TC219353 weakly similar to UP|Q8L555 (Q8L555) Uclacyanin 3-like protein-like
protein, partial (44%)
Length = 713
Score = 62.0 bits (149), Expect = 1e-10
Identities = 41/133 (30%), Positives = 60/133 (44%), Gaps = 4/133 (3%)
Frame = +2
Query: 24 VMGGPKLHKVGGSKGWKENVNYTTWSSQEHVYVGDWLKFVFDKRYYNVLEVNKTGYDYCI 83
++GGP S GW N N +W+S + VGD L F + + +V+EV K YD C
Sbjct: 128 IVGGP-------SGGWDTNSNLQSWASSQIFSVGDSLVFQYPPNH-DVVEVTKADYDSCQ 283
Query: 84 DMTFIRNLTRGGRDVVQLTEAKTYYFITGGGYCFHGMKVAVD----VQEHPTPAPSPSLS 139
I++ G + K Y+ G+C GMKV +D TPA SP S
Sbjct: 284 PTNPIQSYNDGATTIPLTLPGKRYFICGTIGHCSQGMKVEIDTLASATNSVTPAASPEDS 463
Query: 140 DTAKSGGDSILPS 152
T+ + ++ S
Sbjct: 464 TTSPAESPEVIIS 502
>TC207895 similar to UP|O81500 (O81500) F9D12.16 protein (At5g26330), partial
(53%)
Length = 858
Score = 62.0 bits (149), Expect = 1e-10
Identities = 34/112 (30%), Positives = 63/112 (55%), Gaps = 4/112 (3%)
Frame = +3
Query: 30 LHKVGGSKGWK--ENVNYTTWSSQEHVYVGDWLKFVFDKRYYNVLEVNKTGYDYCIDMTF 87
+HKVG S GW N++Y W++ ++ VGD + F ++ +++NV+ V Y C +
Sbjct: 201 VHKVGDSAGWTIIGNIDYKKWAATKNFQVGDTIIFEYNAKFHNVMRVTHAMYKSCNASSP 380
Query: 88 IRNLTRGGRDVVQLTEAKTYYFITG-GGYCFHGMKVAVD-VQEHPTPAPSPS 137
+ ++ G D +++T ++F+ G G+C G KV ++ V+ APSP+
Sbjct: 381 LTTMST-GNDTIKITNYGHHFFLCGIPGHCQAGQKVDINVVKVSAAAAPSPT 533
>TC204674 similar to UP|Q9ZRV5 (Q9ZRV5) Basic blue copper protein, partial
(98%)
Length = 635
Score = 61.2 bits (147), Expect = 2e-10
Identities = 36/115 (31%), Positives = 57/115 (49%)
Frame = +2
Query: 11 SVVLGVLSLWPMVVMGGPKLHKVGGSKGWKENVNYTTWSSQEHVYVGDWLKFVFDKRYYN 70
+VVL + L M ++VG S+GW N TW + GD L F + +N
Sbjct: 74 AVVLLLCFLVLQSEMARAATYRVGDSRGW--TFNTVTWPQGKRFTAGDTLAFNYSPGAHN 247
Query: 71 VLEVNKTGYDYCIDMTFIRNLTRGGRDVVQLTEAKTYYFITGGGYCFHGMKVAVD 125
V+ V+K GYD C + + R G+D ++L + Y+ G+C GMK+A++
Sbjct: 248 VVAVSKAGYDSCKTPRGAK-VYRSGKDQIRLARGQNYFICNYVGHCESGMKIAIN 409
>TC225574 similar to UP|Q9ZRV5 (Q9ZRV5) Basic blue copper protein, partial
(83%)
Length = 868
Score = 60.8 bits (146), Expect = 3e-10
Identities = 36/124 (29%), Positives = 61/124 (49%)
Frame = +1
Query: 2 KNSRVLLMVSVVLGVLSLWPMVVMGGPKLHKVGGSKGWKENVNYTTWSSQEHVYVGDWLK 61
+ S L +V V+ +L L ++ + VGG GW N N W + GD L
Sbjct: 103 RGSASLPIVVTVVSLLCLLVLLERADAATYTVGGPGGWTFNTN--AWPKGKRFRAGDILI 276
Query: 62 FVFDKRYYNVLEVNKTGYDYCIDMTFIRNLTRGGRDVVQLTEAKTYYFITGGGYCFHGMK 121
F +D +NV+ V+++GY+ C + + G+D ++L + Y+ G+C GMK
Sbjct: 277 FNYDSTTHNVVAVDRSGYNSCKTPGGAKVFS-SGKDQIKLARGQNYFICNYPGHCESGMK 453
Query: 122 VAVD 125
VA++
Sbjct: 454 VAIN 465
>TC231303 homologue to UP|N552_SOYBN (Q02917) Early nodulin 55-2 precursor
(N-55-2) (Nodulin-315), complete
Length = 839
Score = 57.0 bits (136), Expect = 4e-09
Identities = 43/141 (30%), Positives = 71/141 (49%), Gaps = 15/141 (10%)
Frame = +2
Query: 11 SVVLGVLSLWPMVVMGGPKLHKVGGS-KGWKENVN----YTTWSSQEHVYVGDWLKFVFD 65
S L +L++ ++ + + VGGS K WK ++ + W++ +GD L F ++
Sbjct: 80 SPFLVMLAMCLLISTSEAEKYVVGGSEKSWKFPLSKPDSLSHWANSHRFKIGDTLIFKYE 259
Query: 66 KRYYNVLEVNKTGYDYCIDMTFIRNLTRGGRDVVQLTEAKTYYFITGG-GYCFHGMKVAV 124
KR +V EVN+T Y+ C + + GG V LT+ +FI+G +C G+K+AV
Sbjct: 260 KRTESVHEVNETDYEGCNTVGKYHIVFNGGNTKVMLTKPGFRHFISGNQSHCQMGLKLAV 439
Query: 125 DVQE---------HPTPAPSP 136
V P+P+PSP
Sbjct: 440 LVISSNKTKKNLLSPSPSPSP 502
>TC209188 similar to UP|BCP_PEA (Q41001) Blue copper protein precursor,
partial (57%)
Length = 597
Score = 56.6 bits (135), Expect = 6e-09
Identities = 43/138 (31%), Positives = 65/138 (46%), Gaps = 6/138 (4%)
Frame = +1
Query: 17 LSLWPMVV--MGGPKL---HKVGGSKGWKENVNYTTWSSQEHVYVGDWLKFVFDKRYYNV 71
L LW ++ M P L + VG + GW +Y+TW+ + VGD L F + + V
Sbjct: 76 LILWSLLAINMALPTLATVYTVGDTSGWAIGTDYSTWTGDKIFSVGDSLAFNYGAGH-TV 252
Query: 72 LEVNKTGYDYCIDMTFIRNLTRGGRDVVQLTEAKTYYFITG-GGYCFHGMKVAVDVQEHP 130
EV ++ Y C I + G + L A T+YFI G+C GMK+AV V+
Sbjct: 253 DEVKESDYKSCTAGNSI-STDSSGATTIALKSAGTHYFICSVPGHCSGGMKLAVTVKSGK 429
Query: 131 TPAPSPSLSDTAKSGGDS 148
+P+ S T + +S
Sbjct: 430 ASDATPTTSTTTATKSNS 483
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.140 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,461,138
Number of Sequences: 63676
Number of extensions: 149475
Number of successful extensions: 966
Number of sequences better than 10.0: 67
Number of HSP's better than 10.0 without gapping: 930
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 944
length of query: 175
length of database: 12,639,632
effective HSP length: 91
effective length of query: 84
effective length of database: 6,845,116
effective search space: 574989744
effective search space used: 574989744
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 55 (25.8 bits)
Medicago: description of AC147482.3


