

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
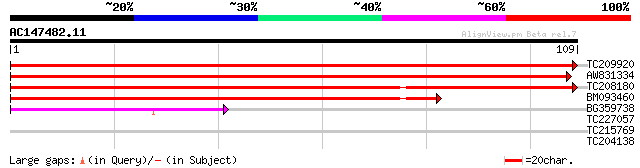
Query= AC147482.11 - phase: 0
(109 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC209920 similar to UP|CHMU_ARATH (P42738) Chorismate mutase, ch... 188 4e-49
AW831334 weakly similar to PIR|S38958|S38 chorismate mutase (EC ... 144 5e-36
TC208180 similar to UP|Q9S7H4 (Q9S7H4) Chorismate mutase CM2 (C... 120 8e-29
BM093460 83 2e-17
BG359738 45 5e-06
TC227057 similar to UP|Q7XB12 (Q7XB12) Homogentisic acid geranyl... 27 1.9
TC215769 similar to GB|AAN28738.1|23505757|AY143799 At2g02050/F1... 25 5.6
TC204138 similar to UP|Q8RVA9 (Q8RVA9) Succinate dehydrogenase s... 25 5.6
>TC209920 similar to UP|CHMU_ARATH (P42738) Chorismate mutase, chloroplast
precursor (CM-1) , partial (52%)
Length = 745
Score = 188 bits (477), Expect = 4e-49
Identities = 91/109 (83%), Positives = 103/109 (94%)
Frame = +2
Query: 1 ALSKRIHYGKFVAEAKFQAAPDSYKAAIIAQDKDRLMELLTYPEVEESIKRRVAVKAKTY 60
ALSKRIHYGK+VAEAK+QA+PDSYK AIIAQDKD+LMELLTYPEVEE+IKRRV +K+KTY
Sbjct: 233 ALSKRIHYGKYVAEAKYQASPDSYKDAIIAQDKDKLMELLTYPEVEEAIKRRVDMKSKTY 412
Query: 61 GQEVAVNLKDQKTEPVYKINPSLVADLYSDWIMPLTKEVQVAYLLRKLD 109
GQE+ V K+ +TEPVYKINPSLVADLYSDWIMPLTKEVQV+YLLR+LD
Sbjct: 413 GQELVVTTKEHRTEPVYKINPSLVADLYSDWIMPLTKEVQVSYLLRRLD 559
>AW831334 weakly similar to PIR|S38958|S38 chorismate mutase (EC 5.4.99.5)
precursor - Arabidopsis thaliana, partial (40%)
Length = 556
Score = 144 bits (364), Expect = 5e-36
Identities = 75/108 (69%), Positives = 86/108 (79%)
Frame = +2
Query: 1 ALSKRIHYGKFVAEAKFQAAPDSYKAAIIAQDKDRLMELLTYPEVEESIKRRVAVKAKTY 60
ALSKRIHYGK+VAEAK+Q++P SY AIIA DKD+LMELLTYPEVEE+I RRV VK KTY
Sbjct: 233 ALSKRIHYGKYVAEAKYQSSPYSYNDAIIAHDKDKLMELLTYPEVEEAITRRVVVKTKTY 412
Query: 61 GQEVAVNLKDQKTEPVYKINPSLVADLYSDWIMPLTKEVQVAYLLRKL 108
QE+ V K+ TEPVYKIN SLVA LYS WIM LTK + Y+L K+
Sbjct: 413 VQELVVTTKEHLTEPVYKINLSLVAYLYSYWIMHLTKGIPCYYVLIKV 556
>TC208180 similar to UP|Q9S7H4 (Q9S7H4) Chorismate mutase CM2 (Chorimate
mutase) , partial (68%)
Length = 1281
Score = 120 bits (302), Expect = 8e-29
Identities = 59/109 (54%), Positives = 81/109 (74%)
Frame = +3
Query: 1 ALSKRIHYGKFVAEAKFQAAPDSYKAAIIAQDKDRLMELLTYPEVEESIKRRVAVKAKTY 60
++S+RIHYGKFVAE KF+ AP Y+ I A+DK+ LM+LLT+ VEE++++RV KA +
Sbjct: 687 SISRRIHYGKFVAEVKFRDAPQDYEPLIRAKDKEGLMKLLTFTSVEETVRKRVEKKAVVF 866
Query: 61 GQEVAVNLKDQKTEPVYKINPSLVADLYSDWIMPLTKEVQVAYLLRKLD 109
GQEV +N D E K +PS+ + LY +W++PLTKEVQV YLLR+LD
Sbjct: 867 GQEVNLNSDDNDNEN-RKFDPSVASSLYKNWVIPLTKEVQVEYLLRRLD 1010
>BM093460
Length = 406
Score = 83.2 bits (204), Expect = 2e-17
Identities = 42/83 (50%), Positives = 59/83 (70%)
Frame = +1
Query: 1 ALSKRIHYGKFVAEAKFQAAPDSYKAAIIAQDKDRLMELLTYPEVEESIKRRVAVKAKTY 60
++S+RIHYGKFVAE KF+ AP Y+ I A+DK+ LM+LLT+ VEE++++RV KA +
Sbjct: 157 SISRRIHYGKFVAEVKFRDAPQDYEPLIRAKDKEGLMKLLTFTSVEETVRKRVEKKAVVF 336
Query: 61 GQEVAVNLKDQKTEPVYKINPSL 83
GQEV +N D E K +PS+
Sbjct: 337 GQEVNLNSDDNDNEN-RKFDPSV 402
>BG359738
Length = 418
Score = 45.1 bits (105), Expect = 5e-06
Identities = 25/64 (39%), Positives = 33/64 (51%), Gaps = 22/64 (34%)
Frame = +1
Query: 1 ALSKRIHYGKFVAEAKFQAAPDSYKA----------------------AIIAQDKDRLME 38
A+S+RIHYGKFV EAKF+ +P Y+ A + +DK LM+
Sbjct: 220 AISRRIHYGKFVGEAKFRESPQGYEPLIRAKIHPNF*YSRKLCTNVAHATVIEDKGALMK 399
Query: 39 LLTY 42
LLTY
Sbjct: 400 LLTY 411
>TC227057 similar to UP|Q7XB12 (Q7XB12) Homogentisic acid geranylgeranyl
transferase, partial (22%)
Length = 1536
Score = 26.6 bits (57), Expect = 1.9
Identities = 12/50 (24%), Positives = 26/50 (52%)
Frame = -3
Query: 49 IKRRVAVKAKTYGQEVAVNLKDQKTEPVYKINPSLVADLYSDWIMPLTKE 98
+ R +++ + + + L+ EPV ++NP L A ++ + P+ KE
Sbjct: 805 LSRGTSIE*AVHDEAITTRLQINGQEPVIQVNPKLRAKNAAEIMTPVLKE 656
>TC215769 similar to GB|AAN28738.1|23505757|AY143799 At2g02050/F14H20.12
{Arabidopsis thaliana;} , partial (76%)
Length = 600
Score = 25.0 bits (53), Expect = 5.6
Identities = 13/48 (27%), Positives = 22/48 (45%)
Frame = +2
Query: 44 EVEESIKRRVAVKAKTYGQEVAVNLKDQKTEPVYKINPSLVADLYSDW 91
EVE S K+ +A + + V + +DQ + +N A+ Y W
Sbjct: 83 EVEGSSKKMIATQEEMVEARVPLAYRDQCAHLLIPLNKCRQAEFYLPW 226
>TC204138 similar to UP|Q8RVA9 (Q8RVA9) Succinate dehydrogenase subunit 4,
partial (74%)
Length = 904
Score = 25.0 bits (53), Expect = 5.6
Identities = 13/48 (27%), Positives = 26/48 (54%)
Frame = +3
Query: 47 ESIKRRVAVKAKTYGQEVAVNLKDQKTEPVYKINPSLVADLYSDWIMP 94
+S ++ A KAK + ++ + ++P+++I+ SLVA S P
Sbjct: 39 DSSHQKKAKKAKMQSMSLTLSKRLSNSKPLFRISSSLVAHASSATTSP 182
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.132 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,248,266
Number of Sequences: 63676
Number of extensions: 25986
Number of successful extensions: 179
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 177
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 177
length of query: 109
length of database: 12,639,632
effective HSP length: 85
effective length of query: 24
effective length of database: 7,227,172
effective search space: 173452128
effective search space used: 173452128
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 51 (24.3 bits)
Medicago: description of AC147482.11


