

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147435.10 + phase: 0 /pseudo
(612 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
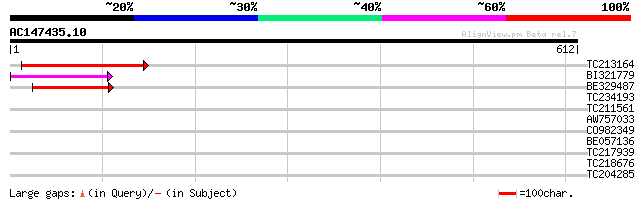
Score E
Sequences producing significant alignments: (bits) Value
TC213164 114 1e-25
BI321779 89 6e-18
BE329487 weakly similar to PIR|T14619|T146 reverse transcriptase... 82 9e-16
TC234193 40 0.004
TC211561 weakly similar to UP|O22675 (O22675) Reverse transcript... 36 0.044
AW757033 35 0.075
CO982349 33 0.28
BE057136 similar to PIR|JQ1060|JQ10 glycine-rich protein 1 - Ara... 30 4.1
TC217939 similar to UP|O49350 (O49350) AT-hook protein 2, partia... 29 5.3
TC218676 weakly similar to UP|Q948Y6 (Q948Y6) VMP4 protein, part... 29 5.3
TC204285 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 29 7.0
>TC213164
Length = 446
Score = 114 bits (285), Expect = 1e-25
Identities = 54/138 (39%), Positives = 89/138 (64%)
Frame = +3
Query: 13 NDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQIHANELL 72
+DLLVA F +E+ + + KSPG D NF F+KEFW ++K ++ D+ +AN +
Sbjct: 9 SDLLVARFEKKEVRATMWDRGNAKSPGPDGLNFKFIKEFWDILKTDLLRFLDEFYANGVF 188
Query: 73 PKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVISTTQSAF 132
K A+F+ALIPKV P L +Y+ ISL+G +YK++AK+L+ R VL ++I Q+ F
Sbjct: 189 LKRSNAFFLALIPKVHDP*SLNEYKPISLIGCIYKIVAKLLSGRHKKVLPTIIDERQTVF 368
Query: 133 LKGTNLVDGVLVVNELVD 150
++G + + V++ NE+++
Sbjct: 369 MEGRHTLHNVVIANEIME 422
>BI321779
Length = 421
Score = 89.0 bits (219), Expect = 6e-18
Identities = 44/111 (39%), Positives = 67/111 (59%)
Frame = +2
Query: 1 DGVNFNRLSEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVR 60
+GV F S DN +L+ F +EEI+ V+ + + +KSPG D Y+ +K FW ++K EV
Sbjct: 38 NGVEFLSFSNEDNAMLIQDFEVEEIKKVI*DCESSKSPGPDGYSLLLIKIFWEVIKEEVI 217
Query: 61 IMFDQIHANELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAK 111
F + H + LP+ F+ALI K +PL++ Y ISL+G LYK++ K
Sbjct: 218 NFFKEFHKFDFLPRGTNRSFIALIAKCDNPLDIGQYCPISLVGCLYKIV*K 370
>BE329487 weakly similar to PIR|T14619|T146 reverse transcriptase - beet
retrotransposon (fragment), partial (15%)
Length = 376
Score = 81.6 bits (200), Expect = 9e-16
Identities = 39/88 (44%), Positives = 55/88 (62%)
Frame = -2
Query: 25 IEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQIHANELLPKSMLAYFVALI 84
I+ V + KSPG D NF F+K FW +K ++ ++ HAN + PK A F+ALI
Sbjct: 375 IKSAVWQCGSVKSPGPDGLNFKFIKHFWERLKPDIIRFLNEFHANGIFPKGGNASFIALI 196
Query: 85 PKVSSPLELKDYRTISLLGSLYKLLAKV 112
PKV P L D+R ISL+G +YK++AK+
Sbjct: 195 PKVKHPQALNDFRPISLIGCVYKIVAKI 112
>TC234193
Length = 697
Score = 39.7 bits (91), Expect = 0.004
Identities = 34/115 (29%), Positives = 55/115 (47%), Gaps = 8/115 (6%)
Frame = -3
Query: 1 DGVNFNRLSEVDNDLLVAPFHMEEIEVVVRESDGNKSPG--------ADEYNFAFVKEFW 52
DGVNF ++ DND L H+ +V+ + S G ++ YNF F+K+
Sbjct: 317 DGVNFKTINGEDNDRLT---HV----LVLGTQNKRGSLGV*WKQKFDSNGYNFNFIKKC* 159
Query: 53 YLMKNEVRIMFDQIHANELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYK 107
+MK +V + H++ L + A F+ P L D+R ++L+GSL K
Sbjct: 158 EVMKEDVVRAIQEFHSHGCLSRGTNASFL-----THPP*GLGDFRPVTLVGSLCK 9
>TC211561 weakly similar to UP|O22675 (O22675) Reverse transcriptase
(Fragment) , partial (59%)
Length = 1077
Score = 36.2 bits (82), Expect = 0.044
Identities = 60/188 (31%), Positives = 81/188 (42%)
Frame = +2
Query: 161 SFSRLILRRHMIP*SGVFWNT**EG*VCVRSGWLG*RLVCLEGVCLFLLMEAQRRRFVFK 220
SFS+ I++RH I G+F * + VR+G G + V + LF M A++ +
Sbjct: 65 SFSKWIMKRHTIRFHGIF*CI*CGEWISVRNG*NGLKSVSNLHLFLF**MVARQLSSYRR 244
Query: 221 EALNKVIL*PRSYFCWLRKGLVV**GMRWI*IYSKVLRWEEMVWWCLICNMPMTLFVLGK 280
L K * Y K +V**G +W I + +MV +M MT F LGK
Sbjct: 245 GVLGKETH*HLFYSILWLKV*MV**GEQWRKICTSPT*LVQMVCPSASYSMLMTQFFLGK 424
Query: 281 LPWRIFGR*RLF*EVLKWLPG*RLIFPKALL*ELIFRRILCKWRAIF*IVVKGVSRSSIW 340
P R+ R L W * +A+L L + K R I IVV + IW
Sbjct: 425 QPRRM*KLLR*SLGHLSWFLI*GSTSLRAVLVCLE*QINGSKRRPIICIVVCWLFHLYIW 604
Query: 341 DYQWGPIR 348
Y GP R
Sbjct: 605 VYP*GPTR 628
>AW757033
Length = 441
Score = 35.4 bits (80), Expect = 0.075
Identities = 29/75 (38%), Positives = 36/75 (47%)
Frame = -3
Query: 237 LRKGLVV**GMRWI*IYSKVLRWEEMVWWCLICNMPMTLFVLGKLPWRIFGR*RLF*EVL 296
L K L+ ** W+ I K W + N PM LF G+ WR+ G *+ F EVL
Sbjct: 370 LLKVLLA**ERLWLEITIKASLWGNIRNPSAFYNTPMILFSSGRQQWRM*GL*KQFSEVL 191
Query: 297 KWLPG*RLIFPKALL 311
L +LI KA L
Sbjct: 190 S*LLVLKLISQKAAL 146
>CO982349
Length = 795
Score = 33.5 bits (75), Expect = 0.28
Identities = 30/94 (31%), Positives = 43/94 (44%), Gaps = 2/94 (2%)
Frame = +1
Query: 194 LG*RLVCLEGVCLFLLMEAQRRRFVFKEALNKVIL*PRSYFCW--LRKGLVV**GMRWI* 251
+G ++ CLF LM+A KEA +K I P + C L+K +V** W
Sbjct: 67 IGSEAASIQHQCLFWLMKALLTNSPHKEASDKAI--PSHHCCLTLLQKV*LV**EKLWTE 240
Query: 252 IYSKVLRWEEMVWWCLICNMPMTLFVLGKLPWRI 285
W ++ + NMPM LF +L W+I
Sbjct: 241 SGLIAS*WGKIRSQLVFYNMPMILFSSKRLQWKI 342
>BE057136 similar to PIR|JQ1060|JQ10 glycine-rich protein 1 - Arabidopsis
thaliana (fragment), partial (19%)
Length = 420
Score = 29.6 bits (65), Expect = 4.1
Identities = 20/51 (39%), Positives = 22/51 (42%)
Frame = -3
Query: 544 LLNLISNGGYGGGLWGDFGGELGLEFYLEAKTICMGGGVAY*PFGRFGGNK 594
L++ GG GGG G FGG G I GGG G FGG K
Sbjct: 148 LVDGFGGGGLGGGAGGGFGGGAG-------AGIGGGGGAGGGAGGGFGGGK 17
>TC217939 similar to UP|O49350 (O49350) AT-hook protein 2, partial (22%)
Length = 542
Score = 29.3 bits (64), Expect = 5.3
Identities = 15/36 (41%), Positives = 19/36 (52%)
Frame = -2
Query: 431 EKWRYRDKIHLANECVFVGKIEMETLRGGNGVVEGG 466
E +R R + LA E F+G+ E GNG EGG
Sbjct: 241 EYFRGRPRFFLAREQAFIGEEEPSYAEAGNGSDEGG 134
>TC218676 weakly similar to UP|Q948Y6 (Q948Y6) VMP4 protein, partial (5%)
Length = 879
Score = 29.3 bits (64), Expect = 5.3
Identities = 20/51 (39%), Positives = 22/51 (42%), Gaps = 4/51 (7%)
Frame = +3
Query: 551 GGYGGGLWGDFGGELG----LEFYLEAKTICMGGGVAY*PFGRFGGNKMVR 597
GGYGGG+ G +GG G F GGGV G GG M R
Sbjct: 213 GGYGGGMGGPYGGYGGGPGSTGFAGAGGMGAGGGGVGGGGLGGAGGGSMYR 365
>TC204285 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (32%)
Length = 1539
Score = 28.9 bits (63), Expect = 7.0
Identities = 18/42 (42%), Positives = 19/42 (44%)
Frame = -2
Query: 551 GGYGGGLWGDFGGELGLEFYLEAKTICMGGGVAY*PFGRFGG 592
GG GGG +G FGG G F GG FG FGG
Sbjct: 635 GGRGGGFFGGFGGSGGTTFGGLGGFFGGFGG*GGSTFGGFGG 510
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.358 0.165 0.605
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 28,256,340
Number of Sequences: 63676
Number of extensions: 435231
Number of successful extensions: 4723
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 2559
Number of HSP's successfully gapped in prelim test: 173
Number of HSP's that attempted gapping in prelim test: 1984
Number of HSP's gapped (non-prelim): 2920
length of query: 612
length of database: 12,639,632
effective HSP length: 103
effective length of query: 509
effective length of database: 6,081,004
effective search space: 3095231036
effective search space used: 3095231036
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC147435.10


