

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147434.6 + phase: 0 /pseudo
(416 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
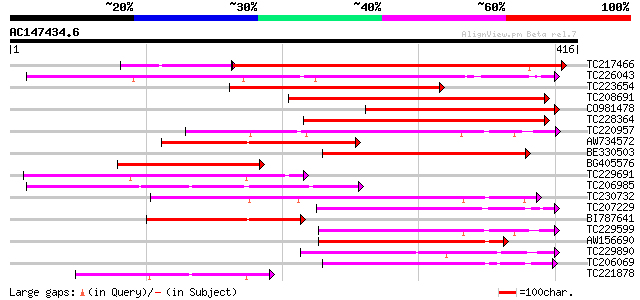
Score E
Sequences producing significant alignments: (bits) Value
TC217466 similar to UP|Q8S9J4 (Q8S9J4) AT5g64600/MUB3_12, partia... 249 5e-83
TC226043 similar to UP|Q9FKP3 (Q9FKP3) Similarity to auxin-indep... 232 2e-61
TC223654 similar to GB|AAD32773.1|4895186|AC007661 axi 1-like pr... 226 1e-59
TC208691 182 2e-46
CO981478 180 9e-46
TC228364 171 6e-43
TC220957 similar to UP|Q9LQE0 (Q9LQE0) F15O4.45, partial (43%) 171 6e-43
AW734572 166 1e-41
BE330503 148 4e-36
BG405576 similar to GP|10176802|db axi 1 (auxin-independent grow... 145 4e-35
TC229691 similar to UP|Q9LIN9 (Q9LIN9) Similarity to the auxin-i... 142 2e-34
TC206985 similar to UP|Q9SSR5 (Q9SSR5) F6D8.15 protein, partial ... 142 2e-34
TC230732 similar to UP|O48796 (O48796) F24O1.5, partial (46%) 136 2e-32
TC207229 homologue to UP|Q9LIN9 (Q9LIN9) Similarity to the auxin... 129 2e-30
BI787641 127 9e-30
TC229599 similar to UP|Q9LQT6 (Q9LQT6) F10B6.36, partial (42%) 120 9e-28
AW156690 118 6e-27
TC229890 similar to GB|AAP68319.1|31711926|BT008880 At1g04910 {A... 110 9e-25
TC206069 similar to UP|O64884 (O64884) Axi 1 protein-like protei... 102 2e-22
TC221878 similar to UP|Q8LR78 (Q8LR78) Axi 1-like protein, parti... 101 5e-22
>TC217466 similar to UP|Q8S9J4 (Q8S9J4) AT5g64600/MUB3_12, partial (84%)
Length = 1326
Score = 249 bits (635), Expect(2) = 5e-83
Identities = 125/254 (49%), Positives = 173/254 (67%), Gaps = 9/254 (3%)
Frame = +1
Query: 164 KKLVNRLRENNTPYIALHLRYEKDMLAFTGCSHNLTKEETQELKKMRYSVKHWKEKEIDS 223
++LV+RLR + YIALHLRYEKDML+FTGC++ LT E++EL+ +R + +WK K+I+S
Sbjct: 277 QRLVDRLRSHGGRYIALHLRYEKDMLSFTGCAYGLTDAESEELRILRENTNYWKVKKINS 456
Query: 224 KSKRLKGSCPMTPREVAVFLEALGYPVDTKIYVAAGVIY-GSEGMKPLQKKFPNLLWHSS 282
+R+ G CP+TP+EV +FL ALGYP T IY+AAGVIY G+ + L +FP++++ S
Sbjct: 457 TEQRIGGFCPLTPKEVGIFLHALGYPPSTPIYIAAGVIYGGNTHLSELSSRFPSIIFKES 636
Query: 283 LATKEELQPFEGHLNQLAALDYYITVESDVFVYSYDGNMAKAARGHRKFDGFKKTISPDK 342
LAT EEL+ F H +Q AALDY I VESDVFV SY GNMA+A GHR+F G +KTI+PD+
Sbjct: 637 LATPEELKDFANHASQTAALDYIICVESDVFVPSYSGNMARAVEGHRRFLGHRKTINPDR 816
Query: 343 QRFVRLIDQLDNG-LISWNDFSTKVKSIHAKKKGAPQAR-------KIHRHPKFEETFYA 394
+ V + D L+ G L+ + S V+ +H ++GAP+ R K + EE FY
Sbjct: 817 KGLVGIFDMLETGELVEGRELSNMVQRMHKNRQGAPRKRHGSLPGIKGRARFRTEEPFYE 996
Query: 395 NPFPGCICQKPTHL 408
NP+P CIC + L
Sbjct: 997 NPYPECICGSKSKL 1038
Score = 77.4 bits (189), Expect(2) = 5e-83
Identities = 42/85 (49%), Positives = 51/85 (59%)
Frame = +3
Query: 82 VVESLPPELAAIKPALKAPVSWSKASYYRTDMLQLLKKHKVIKFTHTDSRLVNNGLASSI 141
+V LP L + A K SWS YY +M +L ++VI +DSRL NN L I
Sbjct: 3 IVSELPKNLEGVPRARKHFTSWSGVGYYE-EMTRLWSDYQVIHVAKSDSRLANNDLPLDI 179
Query: 142 QRVRCRAMYEALRFAVPIEELGKKL 166
QR+RCRAMY ALRF+ PIE LGK L
Sbjct: 180 QRLRCRAMYHALRFSPPIENLGKVL 254
>TC226043 similar to UP|Q9FKP3 (Q9FKP3) Similarity to auxin-independent
growth promoter, partial (79%)
Length = 2154
Score = 232 bits (592), Expect = 2e-61
Identities = 148/430 (34%), Positives = 227/430 (52%), Gaps = 39/430 (9%)
Frame = +2
Query: 13 TNGYLLVHANGGLNQMKTGISDMVAIAKIMNATLVFPTLDHNSFWTDPSDFKEIFNWKNF 72
+ GY+ V +GGLNQ + GI D VA+AKI+NATLV P L+ N W D S F +IF+ +F
Sbjct: 521 SEGYIQVFLDGGLNQQRMGICDAVAVAKILNATLVIPYLELNPVWRDSSSFMDIFDVDHF 700
Query: 73 VEVLNEDVQVVESLPPE----------LAAIKPALKAPVSWSKASYYRTDMLQLLKKHKV 122
++VL +D+ +V+ LP E LA + +KA + A +Y ++L +L+ + +
Sbjct: 701 IDVLKDDISIVKELPKEFSWSTREYYGLAIRETRIKAAPVHASAHWYLENVLPVLQSYGI 880
Query: 123 IKFTHTDSRLVNNGLASSIQRVRCRAMYEALRFAVPIEELGKKLVNRL------------ 170
+ RL + L IQ +RC+ ++AL F I LG L++RL
Sbjct: 881 AAISPFSHRLSFDNLPMDIQHLRCKVNFQALTFVPHIRTLGDALISRLRYPQGSAEEMGS 1060
Query: 171 -----------RENNTPYIALHLRYEKDMLAFTGCSHNLTKEETQELKKMRYSVKHWKEK 219
R+N ++ LHLR++KDM A + C K E L K R + W+ +
Sbjct: 1061NYLQEVTGAGARKNAGKFVVLHLRFDKDMAAHSACDFGGGKAEKLALAKYRQVI--WQGR 1234
Query: 220 EIDS----KSKRLKGSCPMTPREVAVFLEALGYPVDTKIYVAAGVIYGSEG-MKPLQKKF 274
++S + R +G CPMTP EV + L A+G+ T++Y+A+ +YG E + L++ F
Sbjct: 1235VLNSQFTDEELRSQGRCPMTPEEVGLLLAAMGFDNSTRLYLASHKVYGGEARISTLRELF 1414
Query: 275 PNLLWHSSLATKEELQPFEGHLNQLAALDYYITVESDVFVYSYDGNMAKAARGHRKFDGF 334
P + SLA+ EE +G + LAALDYY+ + SD+F+ + GNM A GHR +
Sbjct: 1415PLMEDKKSLASSEERSQIKGKASLLAALDYYVGLHSDIFISASPGNMHNALVGHRTYLNL 1594
Query: 335 KKTISPDKQRFVRLIDQLD-NGLISWNDFSTKVKSIHAKKKGAPQARKIHRHPKFEETFY 393
KTI P+ + L+ QL N I W++F V H ++G + RK PK ++ Y
Sbjct: 1595-KTIRPN----MALMGQLFLNKTIEWSEFQDAVVEGHQNRQGELRLRK----PK--QSIY 1741
Query: 394 ANPFPGCICQ 403
P P C+CQ
Sbjct: 1742TYPAPDCMCQ 1771
>TC223654 similar to GB|AAD32773.1|4895186|AC007661 axi 1-like protein
{Arabidopsis thaliana;} , partial (25%)
Length = 476
Score = 226 bits (576), Expect = 1e-59
Identities = 104/158 (65%), Positives = 127/158 (79%)
Frame = +2
Query: 162 LGKKLVNRLRENNTPYIALHLRYEKDMLAFTGCSHNLTKEETQELKKMRYSVKHWKEKEI 221
LG+ LVNRLR N PYIALHLRYEKDML+FTGCSHNLT EE +EL+ MRY VKHWKEKEI
Sbjct: 2 LGRVLVNRLRNNKEPYIALHLRYEKDMLSFTGCSHNLTAEEAEELRVMRYEVKHWKEKEI 181
Query: 222 DSKSKRLKGSCPMTPREVAVFLEALGYPVDTKIYVAAGVIYGSEGMKPLQKKFPNLLWHS 281
DS +RL+G CPM+PRE A+FL+A+GYP T IY+ AG IYG ++ Q FP + HS
Sbjct: 182 DSVDRRLQGGCPMSPREAAIFLKAMGYPSTTTIYIVAGPIYGGNSLEAFQSVFPKVFSHS 361
Query: 282 SLATKEELQPFEGHLNQLAALDYYITVESDVFVYSYDG 319
+LAT+EEL+PF+ + N+LAALDY + +ESDVFVY+YDG
Sbjct: 362 TLATEEELEPFKPYQNRLAALDYIVALESDVFVYTYDG 475
>TC208691
Length = 928
Score = 182 bits (463), Expect = 2e-46
Identities = 94/194 (48%), Positives = 131/194 (67%), Gaps = 2/194 (1%)
Frame = +1
Query: 205 ELKKMRYSVKHWKEKEIDSKSKRLKGSCPMTPREVAVFLEALGYPVDTKIYVAAGVIYGS 264
EL +MRY+ WKEK I+S KR G CP+TP E A+ L+AL + +IY+AAG IYG
Sbjct: 4 ELTRMRYAYPWWKEKIINSDLKRKDGLCPLTPEETALTLKALDIDQNIQIYIAAGEIYGG 183
Query: 265 EG-MKPLQKKFPNLLWHSSLATKEELQPFEGHLNQLAALDYYITVESDVFVYSYDGNMAK 323
E M L K++P L+ +L +LQ F+ H +Q+AALDY +++ESD+FV +YDGNMAK
Sbjct: 184 ERRMASLAKEYPKLVRKETLLEPSDLQFFQNHSSQMAALDYLVSLESDIFVPTYDGNMAK 363
Query: 324 AARGHRKFDGFKKTISPDKQRFVRLIDQLDNGLISWNDFSTKVKSIHAKKKGAPQAR-KI 382
GHR++ GFKKTI +++ V LIDQ +NG+++W++FS+ VK HA + G+ R I
Sbjct: 364 VVEGHRRYLGFKKTILLNRKLLVELIDQYNNGVLNWDEFSSAVKEAHANRMGSQTKRFVI 543
Query: 383 HRHPKFEETFYANP 396
PK E+ FYANP
Sbjct: 544 PDKPKEEDYFYANP 585
>CO981478
Length = 818
Score = 180 bits (457), Expect = 9e-46
Identities = 80/142 (56%), Positives = 102/142 (71%)
Frame = -2
Query: 262 YGSEGMKPLQKKFPNLLWHSSLATKEELQPFEGHLNQLAALDYYITVESDVFVYSYDGNM 321
YG MK + FPN+ HSSL +EEL F+ H N LA +DY + ++SDVF+Y+YDGNM
Sbjct: 811 YGRGSMKXXEDDFPNIFSHSSLXXEEELNSFKNHQNMLAGIDYVVALKSDVFLYTYDGNM 632
Query: 322 AKAARGHRKFDGFKKTISPDKQRFVRLIDQLDNGLISWNDFSTKVKSIHAKKKGAPQARK 381
AKA +GHR+F+ F KTI+PDK FV+L+DQLD G ISW FS+KVK +H + GAP R+
Sbjct: 631 AKAVQGHRRFENFMKTINPDKMNFVKLVDQLDEGKISWKKFSSKVKKLHTDRIGAPYPRE 452
Query: 382 IHRHPKFEETFYANPFPGCICQ 403
PK EE+FYANP PGCIC+
Sbjct: 451 TGEFPKLEESFYANPLPGCICE 386
>TC228364
Length = 1129
Score = 171 bits (433), Expect = 6e-43
Identities = 87/183 (47%), Positives = 121/183 (65%), Gaps = 2/183 (1%)
Frame = +3
Query: 216 WKEKEIDSKSKRLKGSCPMTPREVAVFLEALGYPVDTKIYVAAGVIYGSEG-MKPLQKKF 274
WKEK I+S KR G CP+TP E A+ L AL + +IY+AAG IYG + M L K +
Sbjct: 15 WKEKIINSDLKRKDGLCPLTPEETALTLRALDIDQNIQIYIAAGEIYGGDRRMASLAKNY 194
Query: 275 PNLLWHSSLATKEELQPFEGHLNQLAALDYYITVESDVFVYSYDGNMAKAARGHRKFDGF 334
P L+ +L +LQ F+ H +Q+AALDY +++ESD+FV +YDGNMAK GHR++ GF
Sbjct: 195 PKLVRKETLLEPSDLQFFQNHSSQMAALDYLVSLESDIFVPTYDGNMAKVVEGHRRYLGF 374
Query: 335 KKTISPDKQRFVRLIDQLDNGLISWNDFSTKVKSIHAKKKGAPQARKI-HRHPKFEETFY 393
KKTI +++ V LIDQ +G+++WN+FS+ VK +HA + G R + PK E+ FY
Sbjct: 375 KKTILLNRKLLVDLIDQYHDGILNWNEFSSAVKEVHADRMGGATKRLVMPDRPKEEDYFY 554
Query: 394 ANP 396
ANP
Sbjct: 555 ANP 563
>TC220957 similar to UP|Q9LQE0 (Q9LQE0) F15O4.45, partial (43%)
Length = 1035
Score = 171 bits (433), Expect = 6e-43
Identities = 106/290 (36%), Positives = 160/290 (54%), Gaps = 15/290 (5%)
Frame = +1
Query: 130 SRLVNNGLASSIQRVRCRAMYEALRFAVPIEELGKKLVNRLRENNT----PYIALHLRYE 185
S + + S IQ +RC A + ALRF+ PI L + +V+R+ + ++ Y+++HLR+E
Sbjct: 91 SNRLAQAVPSKIQGLRCFANFGALRFSEPIRTLAESMVDRMVKYSSHSGGKYVSVHLRFE 270
Query: 186 KDMLAFTGCSHNLTKEETQELKKMRYSVKHW------KEKEIDSKSKRLKGSCPMTPREV 239
+DM+AF+ C ++ +EE E+ R + W K + I + R+ G CP+TP EV
Sbjct: 271 EDMVAFSCCEYDGGEEEKHEMDIARE--RSWRGKFRRKHRIIKPGANRVDGRCPLTPLEV 444
Query: 240 AVFLEALGYPVDTKIYVAAGVIYGSEG-MKPLQKKFPNLLWHSSLATKEELQPFEGHLNQ 298
+ L +G+ T +YVAAG IY + M PL++ FP L ++LAT EEL F GH +
Sbjct: 445 GMMLRGMGFDNTTSVYVAAGKIYKEQKYMAPLKQMFPRLQTKNTLATPEELAQFMGHSTR 624
Query: 299 LAALDYYITVESDVFVYSYDGNMAKAARGHRK--FDGFKKTISPDKQRFVRLIDQLDNGL 356
LAALDY + + S+VFV + GN GHR+ + G KTI PDK+R L DN
Sbjct: 625 LAALDYTVCLHSEVFVTTQGGNFPHFLMGHRRYMYGGHAKTIKPDKRRLALL---FDNPN 795
Query: 357 ISWNDFSTKVKSI--HAKKKGAPQARKIHRHPKFEETFYANPFPGCICQK 404
I W F ++K + H+ +KG K E+ Y P P C+C++
Sbjct: 796 IRWEVFKQQMKDMLRHSDQKGT-------ELKKAGESLYTFPMPDCMCRQ 924
>AW734572
Length = 443
Score = 166 bits (421), Expect = 1e-41
Identities = 80/146 (54%), Positives = 109/146 (73%)
Frame = +2
Query: 112 DMLQLLKKHKVIKFTHTDSRLVNNGLASSIQRVRCRAMYEALRFAVPIEELGKKLVNRLR 171
++ L + ++VI+ + +DSRL NN L IQ++RCRA YEALRF+ IE++GK LV R+R
Sbjct: 8 EIASLWEDYQVIRASKSDSRLANNNLPPDIQKLRCRACYEALRFSPRIEQMGKLLVERMR 187
Query: 172 ENNTPYIALHLRYEKDMLAFTGCSHNLTKEETQELKKMRYSVKHWKEKEIDSKSKRLKGS 231
PYIALHLRYEKDMLAF+GC+H+L+ E +EL+ +R ++ +WK KEID +R KG
Sbjct: 188 SFG-PYIALHLRYEKDMLAFSGCTHDLSPVEAEELRSIRENISYWKIKEIDPIEQRSKGL 364
Query: 232 CPMTPREVAVFLEALGYPVDTKIYVA 257
CP+TP+EV +FL ALGYP T IY+A
Sbjct: 365 CPLTPKEVGIFLTALGYPSTTPIYIA 442
>BE330503
Length = 491
Score = 148 bits (374), Expect = 4e-36
Identities = 73/154 (47%), Positives = 100/154 (64%), Gaps = 1/154 (0%)
Frame = +3
Query: 230 GSCPMTPREVAVFLEALGYPVDTKIYVAAGVIYGSEG-MKPLQKKFPNLLWHSSLATKEE 288
G CP+TP E + L ALG + +IY+AAG IYG + M LQ FPNL+ +L +
Sbjct: 18 GLCPLTPEETTLILTALGIDQNIQIYIAAGEIYGGQRRMASLQAAFPNLVRKETLLEPSD 197
Query: 289 LQPFEGHLNQLAALDYYITVESDVFVYSYDGNMAKAARGHRKFDGFKKTISPDKQRFVRL 348
L F+ H +Q+AALDY +++ESD+F+ +YDGNMAK GHR+F GFK+TI D++ V L
Sbjct: 198 LMYFQNHSSQMAALDYLVSLESDIFIPTYDGNMAKVVEGHRRFLGFKRTILLDRKHLVHL 377
Query: 349 IDQLDNGLISWNDFSTKVKSIHAKKKGAPQARKI 382
ID G +SW++FS VK HA + G P+ R I
Sbjct: 378 IDLYTKGSLSWDEFSIMVKKSHANRMGNPKRRVI 479
>BG405576 similar to GP|10176802|db axi 1 (auxin-independent growth
promoter)-like protein {Arabidopsis thaliana}, partial
(16%)
Length = 327
Score = 145 bits (365), Expect = 4e-35
Identities = 65/108 (60%), Positives = 85/108 (78%)
Frame = +2
Query: 80 VQVVESLPPELAAIKPALKAPVSWSKASYYRTDMLQLLKKHKVIKFTHTDSRLVNNGLAS 139
V +VE LPP A I+P K P+SWSK YY+T++L LLK+HKV+ FTHTDSRL NN +
Sbjct: 2 VHIVEKLPPAYAGIEPFPKTPISWSKVPYYKTEVLPLLKQHKVMYFTHTDSRLDNNDIPR 181
Query: 140 SIQRVRCRAMYEALRFAVPIEELGKKLVNRLRENNTPYIALHLRYEKD 187
SIQ++RCRA Y AL+++ P+EELG LV+R+++N PY+ALHLRYEKD
Sbjct: 182 SIQKLRCRANYRALKYSAPVEELGNTLVSRMQQNGNPYLALHLRYEKD 325
>TC229691 similar to UP|Q9LIN9 (Q9LIN9) Similarity to the auxin-independent
growth promoter, partial (56%)
Length = 1392
Score = 142 bits (359), Expect = 2e-34
Identities = 76/219 (34%), Positives = 125/219 (56%), Gaps = 10/219 (4%)
Frame = +2
Query: 11 NNTNGYLLVHANGGLNQMKTGISDMVAIAKIMNATLVFPTLDHNSFWTDPSDFKEIFNWK 70
N TNGY+ +HA GGLNQ + I + VA+AKI+NATL+ P L + W D + F++IF+
Sbjct: 725 NETNGYIFIHAEGGLNQQRIAICNAVAVAKILNATLILPVLKQDQIWKDQTKFEDIFDVD 904
Query: 71 NFVEVLNEDVQVVESLP------PEL-AAIKPALKAPVSWSKASYYRTDMLQLLKKHKVI 123
+F++ L DV++V +P EL +I+ +K ++ A +Y ++L +K+ K++
Sbjct: 905 HFIDYLKYDVRIVRDIPTWFTDKSELFTSIRRTVKNIPKYAPAQFYIDNVLPRVKEKKIM 1084
Query: 124 KFTHTDSRLVNNGLASSIQRVRCRAMYEALRFAVPIEELGKKLVNRLRE---NNTPYIAL 180
RL + + I ++RCR Y AL+F IE++ L +R+R ++ PY+AL
Sbjct: 1085ALKPFVDRLGYDNVPPEINKLRCRVNYHALKFLPDIEQMANSLASRMRNRTGSSNPYMAL 1264
Query: 181 HLRYEKDMLAFTGCSHNLTKEETQELKKMRYSVKHWKEK 219
HLR+EK M+ + C T++E + K Y K W +
Sbjct: 1265HLRFEKGMVGLSFCDFVGTRDE--KAKMAEYRKKEWPRR 1375
>TC206985 similar to UP|Q9SSR5 (Q9SSR5) F6D8.15 protein, partial (64%)
Length = 1050
Score = 142 bits (359), Expect = 2e-34
Identities = 84/249 (33%), Positives = 144/249 (57%), Gaps = 2/249 (0%)
Frame = +2
Query: 13 TNGYLLVHANGGLNQMKTGISDMVAIAKIMNATLVFPTLDHNSFWTDPSDFKEIFNWKNF 72
T GY+ V GGLNQM+ D V IA+++NATLV P + S+W + S F ++++ F
Sbjct: 269 TYGYIRVDCYGGLNQMRRDFCDGVGIARLLNATLVLPKFEVASYWNETSGFADVYDVDYF 448
Query: 73 VEVLNEDVQVVESLPPELAAIKPALKAPVSWSKASY-YRTDMLQLLKKHKVIKFTHTDSR 131
++ +N V+VV+ LPP++A+ +P ++ S K + Y +L L KHK I T S+
Sbjct: 449 IKHMNGFVKVVKELPPDIASKEP-VRVDCSKRKGQFDYVESVLPSLLKHKYISITPAMSQ 625
Query: 132 LVNNGLASSIQRVRCRAMYEALRFAVPIEELGKKLVNRLRENNTPYIALHLRYEKDMLAF 191
+ + C+A Y+ALR +E +L++ + + P+++LHLR+E DM+A+
Sbjct: 626 R-RDRYPLYAKAALCQACYKALRLTRSLEMKASQLLDAIPK---PFLSLHLRFEPDMVAY 793
Query: 192 TGCSH-NLTKEETQELKKMRYSVKHWKEKEIDSKSKRLKGSCPMTPREVAVFLEALGYPV 250
+ C + +L+ + ++ + K W + ++ RL+G CP+TP E A+ L++L P+
Sbjct: 794 SQCEYPDLSPASIKAIEAAQVDRKPWTGEL--ARVWRLRGKCPLTPNETALILQSLSIPL 967
Query: 251 DTKIYVAAG 259
T IY+AAG
Sbjct: 968 TTNIYLAAG 994
>TC230732 similar to UP|O48796 (O48796) F24O1.5, partial (46%)
Length = 1178
Score = 136 bits (342), Expect = 2e-32
Identities = 98/329 (29%), Positives = 159/329 (47%), Gaps = 42/329 (12%)
Frame = +2
Query: 104 SKASYYRTDMLQLLKKHKVIKFTHTDSRLVNNGLASSIQRVRCRAMYEALRFAVPIEELG 163
+K S+Y +L ++ K++V+ F +RL + +A +QR RCR + AL+F I+E G
Sbjct: 20 AKPSFYLKHILPIILKNQVVHFVGFGNRLAFDPIAFELQRFRCRCNFHALQFVPRIQETG 199
Query: 164 KKLVNRLRENN-----------------------------TPYIALHLRYEKDMLAFTGC 194
L+ RLRE++ + Y+ALHLR+E DM+A + C
Sbjct: 200 ALLLKRLREHSGLIGPLDRYLVGPFAESMKEKSESNAKKASKYLALHLRFEIDMVAHSLC 379
Query: 195 SHNLTKEETQELKKMR------YSVKHWKEKEIDSKSKRLKGSCPMTPREVAVFLEALGY 248
+EE +EL+ R S+ K R +G CP+TP E + L ALG+
Sbjct: 380 EFAGGEEERKELEAYREIHFPALSLLKRTTKLPSPSELRSEGLCPLTPEESILMLGALGF 559
Query: 249 PVDTKIYVAAGVIY-GSEGMKPLQKKFPNLLWHSSLATKEELQPFEGHLNQLAALDYYIT 307
T I+VA +Y G + L +P L+ +L + EL+ F + +QLAALD+
Sbjct: 560 NRKTHIFVAGSNLYGGGSRLVALTNLYPKLVTKENLLSSAELKSFANYSSQLAALDFIGC 739
Query: 308 VESDVFVYSYDGN-MAKAARGHRKF--DGFKKTISPDKQRFVRLIDQLDNGLISWNDFST 364
SD F + G+ ++ G+R + G TI P+K+R + + N I W F
Sbjct: 740 TASDAFAMTDSGSQLSSLVSGYRIYYGGGRMPTIRPNKRRLASIF--MKNSTIEWRVFEQ 913
Query: 365 KVKSIHAKKKGA---PQARKIHRHPKFEE 390
+V+ + K P+AR ++R+P+ +E
Sbjct: 914 RVRKAVRQTKHVQTRPKARSVYRYPRCKE 1000
>TC207229 homologue to UP|Q9LIN9 (Q9LIN9) Similarity to the auxin-independent
growth promoter, partial (34%)
Length = 1091
Score = 129 bits (324), Expect = 2e-30
Identities = 72/180 (40%), Positives = 103/180 (57%), Gaps = 2/180 (1%)
Frame = +1
Query: 226 KRLKGSCPMTPREVAVFLEALGYPVDTKIYVAAGVIYGSEG-MKPLQKKFPNLLWHSSLA 284
KR +G CP+ P EVAV L A+GYP +T+IYVA+G +YG + M PL+ FPNL+ LA
Sbjct: 55 KRKEGRCPLEPGEVAVILRAMGYPKETQIYVASGQVYGGQNRMAPLRNMFPNLVTKEELA 234
Query: 285 TKEELQPFEGHLNQLAALDYYITVESDVFVYSYDGNMAKAARGHRKFDGFK-KTISPDKQ 343
TKEEL F H+ LAALD+ + ++SDVFV ++ GN AK G R++ G + K+I PDK
Sbjct: 235 TKEELDGFRKHVTSLAALDFLVCLKSDVFVMTHGGNFAKLIIGARRYMGHRLKSIKPDKG 414
Query: 344 RFVRLIDQLDNGLISWNDFSTKVKSIHAKKKGAPQARKIHRHPKFEETFYANPFPGCICQ 403
+ + + W F V H + G P+ P ++ + NP C+C+
Sbjct: 415 LMSK---SFGDPYMGWAPFVEDVVVTHQTRTGLPE----ETFPNYD--LWENPLTPCMCR 567
>BI787641
Length = 349
Score = 127 bits (319), Expect = 9e-30
Identities = 58/117 (49%), Positives = 80/117 (67%)
Frame = +1
Query: 101 VSWSKASYYRTDMLQLLKKHKVIKFTHTDSRLVNNGLASSIQRVRCRAMYEALRFAVPIE 160
+SWS SYY+ +L L++K+KV+ TD+RL NN IQR+RCR + ALRF IE
Sbjct: 1 ISWSDISYYKNQILPLIQKYKVVHLNRTDARLANNDQPLEIQRLRCRVNFSALRFTSQIE 180
Query: 161 ELGKKLVNRLRENNTPYIALHLRYEKDMLAFTGCSHNLTKEETQELKKMRYSVKHWK 217
ELGK+++ LR+N P++ LHLRYE DMLAF+GC+ +E +EL +MRY+ WK
Sbjct: 181 ELGKRVIKLLRQNG-PFLVLHLRYEMDMLAFSGCTQGCNSDEVEELTRMRYAYPWWK 348
>TC229599 similar to UP|Q9LQT6 (Q9LQT6) F10B6.36, partial (42%)
Length = 1320
Score = 120 bits (302), Expect = 9e-28
Identities = 67/182 (36%), Positives = 104/182 (56%), Gaps = 5/182 (2%)
Frame = +3
Query: 227 RLKGSCPMTPREVAVFLEALGYPVDTKIYVAAGVIYGSEG-MKPLQKKFPNLLWHSSLAT 285
R+ G CP+TP EV + L +G+ +T I++A+G IY +E M PL + FPNL +LA+
Sbjct: 81 RINGKCPLTPLEVGLMLRGMGFTKNTSIFLASGKIYNAEKTMAPLLQMFPNLHTKETLAS 260
Query: 286 KEELQPFEGHLNQLAALDYYITVESDVFVYSYDGNMAKAARGHRKF--DGFKKTISPDKQ 343
+EEL PF+ + +++AA+DY + ++S+VFV + GN GHR+F G KTI PDK+
Sbjct: 261 EEELAPFKNYSSRMAAIDYTVCLQSEVFVTTQGGNFPHFLLGHRRFLYGGHAKTIKPDKR 440
Query: 344 RFVRLIDQLDNGLISWNDFSTKVKSI--HAKKKGAPQARKIHRHPKFEETFYANPFPGCI 401
+ L DN I W ++ S+ H+ KG R ++ Y+ P P C+
Sbjct: 441 KLALL---FDNPNIGWKSLKRQLLSMRSHSDSKGVELKRP-------NDSIYSFPCPDCM 590
Query: 402 CQ 403
C+
Sbjct: 591 CR 596
>AW156690
Length = 429
Score = 118 bits (295), Expect = 6e-27
Identities = 62/141 (43%), Positives = 89/141 (62%), Gaps = 1/141 (0%)
Frame = +1
Query: 227 RLKGSCPMTPREVAVFLEALGYPVDTKIYVAAGVIYGSEG-MKPLQKKFPNLLWHSSLAT 285
R +G CP+TP EV + L ALGY D IYVA+G +YG + + PL+ FPN ++AT
Sbjct: 13 RRQGRCPLTPEEVGLMLRALGYGSDIHIYVASGEVYGGKRTLAPLRALFPNFHSKETIAT 192
Query: 286 KEELQPFEGHLNQLAALDYYITVESDVFVYSYDGNMAKAARGHRKFDGFKKTISPDKQRF 345
KEEL+PF +++AALD+ + ESDVFV + +GNMAK G R++ G K TI P+ ++
Sbjct: 193 KEELEPFSSFSSRMAALDFIVCDESDVFVTNNNGNMAKILAGRRRYFGHKPTIRPNAKKL 372
Query: 346 VRLIDQLDNGLISWNDFSTKV 366
RL L+ +W F++ V
Sbjct: 373 YRLF--LNRSNSTWEAFASXV 429
>TC229890 similar to GB|AAP68319.1|31711926|BT008880 At1g04910 {Arabidopsis
thaliana;} , partial (39%)
Length = 1153
Score = 110 bits (276), Expect = 9e-25
Identities = 69/194 (35%), Positives = 103/194 (52%), Gaps = 4/194 (2%)
Frame = +1
Query: 214 KHWKEKEIDSKSKRLKGSCPMTPREVAVFLEALGYPVDTKIYVAAGVIYGSEG-MKPLQK 272
+++ K + +R G CP+TP+EV + L ALG+ T+IY+AAG ++G + MKP +
Sbjct: 19 ENFAPKRLVYNERRAIGKCPLTPQEVGLILRALGFDNSTRIYLAAGELFGGDRFMKPFRS 198
Query: 273 KFPNLLWHSSLATKEELQPFEGHLNQLAALDYYITVESDVFVYSYDG--NMAKAARGHRK 330
FP L HSS+ EEL L +A+DY + + SD+F+ +YDG N A GHR
Sbjct: 199 LFPRLENHSSVENSEELAENTRGLAG-SAVDYMVCLLSDIFMPTYDGPSNFANNLLGHRL 375
Query: 331 FDGFKKTISPDKQRFVRLIDQLDNGLISWNDFSTKVKSIHAKKK-GAPQARKIHRHPKFE 389
+ GF+ TI PD++ + +NG + F V+ + K G P R
Sbjct: 376 YYGFRTTIRPDRKSLAPIFIDRENGQTA--GFEEAVRKVMLKTNFGEPHKR------VSP 531
Query: 390 ETFYANPFPGCICQ 403
E+FY N +P C CQ
Sbjct: 532 ESFYTNSWPECFCQ 573
>TC206069 similar to UP|O64884 (O64884) Axi 1 protein-like protein, partial
(30%)
Length = 514
Score = 102 bits (255), Expect = 2e-22
Identities = 62/174 (35%), Positives = 94/174 (53%), Gaps = 1/174 (0%)
Frame = +1
Query: 230 GSCPMTPREVAVFLEALGYPVDTKIYVAAGV-IYGSEGMKPLQKKFPNLLWHSSLATKEE 288
G CP+ EV L+ LG P + +IY A G + G E ++PL +FP+L LA E
Sbjct: 4 GLCPLNSIEVTRLLKGLGAPKNARIYWAGGQPLGGKEVLQPLINEFPHLYSKEDLALHGE 183
Query: 289 LQPFEGHLNQLAALDYYITVESDVFVYSYDGNMAKAARGHRKFDGFKKTISPDKQRFVRL 348
L+PF + +AA+DY ++ +SDVF+ S+ GNM A +GHR + G KK I+P+K++ +
Sbjct: 184 LEPFANKASLMAAIDYIVSEKSDVFMPSHGGNMGHALQGHRAYAGHKKYITPNKRQMLPY 363
Query: 349 IDQLDNGLISWNDFSTKVKSIHAKKKGAPQARKIHRHPKFEETFYANPFPGCIC 402
LD+ L +F+ +K +H G P+ R K P P C+C
Sbjct: 364 F--LDSSLPE-EEFNRIIKELHQDSLGQPE----FRTSKSGRDVTKYPVPECMC 504
>TC221878 similar to UP|Q8LR78 (Q8LR78) Axi 1-like protein, partial (29%)
Length = 507
Score = 101 bits (252), Expect = 5e-22
Identities = 60/156 (38%), Positives = 91/156 (57%), Gaps = 10/156 (6%)
Frame = +2
Query: 49 PTLDHNSFWTDPSDFKEIFNWKNFVEVLNEDVQVVESLPPELAAIKPALKAPV------S 102
P L +S W DPS F++I++ + FV L DV+VV+ +P L + V +
Sbjct: 32 PQLSLHSIWKDPSKFQDIYDEEFFVNTLKNDVRVVDKIPEYLMERFGSNMTNVHNFRIKA 211
Query: 103 WSKASYYRTDMLQLLKKHKVIKFTHTDSRLVNNGLASSIQRVRCRAMYEALRFAVPIEEL 162
WS YYR +L L + KVI+ + +RL + S +Q +RC A YEALRF+ PI +
Sbjct: 212 WSSIQYYRDVVLPKLLEEKVIRISPFANRL-SFDAPSVVQHLRCLANYEALRFSSPILTI 388
Query: 163 GKKLVNRLRE----NNTPYIALHLRYEKDMLAFTGC 194
G+ LV R+R+ N Y+++HLR+E+DM+AF+ C
Sbjct: 389 GESLVERMRKHSAINGGKYVSVHLRFEEDMVAFSCC 496
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,463,186
Number of Sequences: 63676
Number of extensions: 237834
Number of successful extensions: 1256
Number of sequences better than 10.0: 86
Number of HSP's better than 10.0 without gapping: 1197
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1206
length of query: 416
length of database: 12,639,632
effective HSP length: 100
effective length of query: 316
effective length of database: 6,272,032
effective search space: 1981962112
effective search space used: 1981962112
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC147434.6


