

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147431.1 + phase: 0 /pseudo
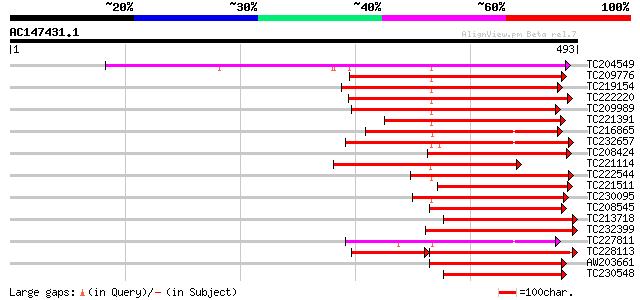
(493 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204549 similar to UP|Q9M0X5 (Q9M0X5) Receptor protein kinase-l... 325 4e-89
TC209776 similar to UP|O65470 (O65470) Serine/threonine kinase-l... 251 5e-67
TC219154 similar to PIR|T05754|T05754 S-receptor kinase M4I22.1... 243 1e-64
TC222220 weakly similar to UP|Q9ZP16 (Q9ZP16) Receptor-like prot... 237 1e-62
TC209989 similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kin... 223 2e-58
TC221391 similar to UP|O81906 (O81906) Serine/threonine kinase-l... 211 8e-55
TC216865 similar to UP|Q9LPG0 (Q9LPG0) T3F20.24 protein, partial... 191 5e-49
TC232657 weakly similar to UP|O22580 (O22580) Receptor-like seri... 190 1e-48
TC208424 weakly similar to UP|O65466 (O65466) Serine/threonine k... 181 5e-46
TC221114 similar to UP|Q8RZ15 (Q8RZ15) Receptor kinase-like prot... 177 7e-45
TC222544 weakly similar to UP|O23743 (O23743) SFR1 protein precu... 174 8e-44
TC221511 similar to UP|Q40096 (Q40096) Receptor protein kinase, ... 170 2e-42
TC230095 weakly similar to UP|Q9XED4 (Q9XED4) Receptor-like prot... 168 4e-42
TC208545 weakly similar to UP|Q9S972 (Q9S972) ARK2 product/recep... 168 6e-42
TC213718 similar to UP|Q70I29 (Q70I29) S-receptor kinase-like pr... 167 1e-41
TC232399 similar to UP|Q39086 (Q39086) Receptor kinase , partial... 167 1e-41
TC227811 similar to PIR|JQ1674|JQ1674 protein kinase TMK1 , rece... 166 3e-41
TC228113 similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kin... 165 4e-41
AW203661 162 2e-40
TC230548 weakly similar to UP|Q8LLI4 (Q8LLI4) S-locus receptor-l... 161 7e-40
>TC204549 similar to UP|Q9M0X5 (Q9M0X5) Receptor protein kinase-like protein,
partial (54%)
Length = 2032
Score = 325 bits (832), Expect = 4e-89
Identities = 191/476 (40%), Positives = 248/476 (51%), Gaps = 72/476 (15%)
Frame = +3
Query: 84 DVVYGFYDCRGDVTGSFCQFCINAAVRAIAQCCPNGVSAMIWYDICVIGYSDHNTSGKIS 143
D VYG + CRGDV + CQ C+ A + C A+IWYD C + YS+ + +
Sbjct: 27 DRVYGLFMCRGDVPSALCQQCVVNATGRLRSQCSLAKQAVIWYDECTVRYSNRSFFSTVD 206
Query: 144 VTPSWNLTGTKNTKDSTELEKAVDDMRNLIGRVTAEAN---SNWAVGEFDWSDTEKRYGW 200
P L T N + + + N A + +AV + + S + Y
Sbjct: 207 TRPRVGLLNTANISNQDSFMRLLFQTINRTADEAANFSVGLKKYAVNQANISGFQSLYCL 386
Query: 201 VQCNSDLTKDGCRYCLETMLDKVPQCCGTKVKWAVVSPSCGMEIDDNKFYNFQTESPPSL 260
QC DL+++ CR CL ++ +P CC K V+ PSC + + FY P S
Sbjct: 387 AQCTPDLSQENCRSCLSGVIGDLPWCCQGKQGGRVLYPSCNVRYELYPFYRVTASPPSSS 566
Query: 261 PNPGGLLLRTITPKSFRDH-----------VP---------------------------R 282
P+P LL +P S VP +
Sbjct: 567 PSPPTLLPPPTSPISPGSSGISAGTIVAIVVPITVAVLIFIVGICFLSRRARKKQQGSVK 746
Query: 283 EDSFNGDLPTIP-----LTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSE 337
E D+PT+ + I+ +T+ FS KLGEGGFG VYKGTL G +A KRLS+
Sbjct: 747 EGKTAYDIPTVDSLQFDFSTIEAATNKFSADNKLGEGGFGEVYKGTLSSGQVVAVKRLSK 926
Query: 338 TSGQGLEEFKNEVIFIAKLQHRNLVKLL--------------------------DEEKHK 371
+SGQG EEFKNEV+ +AKLQHRNLV+LL D EK +
Sbjct: 927 SSGQGGEEFKNEVVVVAKLQHRNLVRLLGFCLQGEEKILVYEYVPNKSLDYILFDPEKQR 1106
Query: 372 HLDWKLRLSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEK 431
LDW R II GIARG+ YLHEDS LR+IHRDLKASN+LLD +MNPKISDFG+AR F
Sbjct: 1107ELDWGRRYKIIGGIARGIQYLHEDSRLRIIHRDLKASNILLDGDMNPKISDFGMARIFGV 1286
Query: 432 DQCHTKTKRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSE 487
DQ T R++GTYGYMAPEYAM G FSVKSDV+SFGVL++EI+ GK+N F+ ++
Sbjct: 1287DQTQGNTSRIVGTYGYMAPEYAMHGEFSVKSDVYSFGVLLMEILSGKKNSSFYQTD 1454
>TC209776 similar to UP|O65470 (O65470) Serine/threonine kinase-like protein
, partial (47%)
Length = 1305
Score = 251 bits (641), Expect = 5e-67
Identities = 129/215 (60%), Positives = 151/215 (70%), Gaps = 26/215 (12%)
Frame = +2
Query: 296 TVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQGLEEFKNEVIFIAK 355
+ I+ +T FSE+ KLGEGGFG VYKG LP G E+A KRLS+ SGQG EEFKNEV +AK
Sbjct: 53 STIEAATQKFSEANKLGEGGFGEVYKGLLPSGQEVAVKRLSKISGQGGEEFKNEVEIVAK 232
Query: 356 LQHRNLVKLL--------------------------DEEKHKHLDWKLRLSIIKGIARGL 389
LQHRNLV+LL D EK K LDW R I++GIARG+
Sbjct: 233 LQHRNLVRLLGFCLEGEEKILVYEFVANKSLDYILFDPEKQKSLDWTRRYKIVEGIARGI 412
Query: 390 LYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKRVIGTYGYMA 449
YLHEDS L++IHRDLKASNVLLD +MNPKISDFG+AR F DQ T R++GTYGYM+
Sbjct: 413 QYLHEDSRLKIIHRDLKASNVLLDGDMNPKISDFGMARIFGVDQTQANTNRIVGTYGYMS 592
Query: 450 PEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFF 484
PEYAM G +S KSDV+SFGVL+LEII GKRN F+
Sbjct: 593 PEYAMHGEYSAKSDVYSFGVLILEIISGKRNSSFY 697
>TC219154 similar to PIR|T05754|T05754 S-receptor kinase M4I22.110 precursor
- Arabidopsis thaliana {Arabidopsis thaliana;} ,
partial (31%)
Length = 1363
Score = 243 bits (620), Expect = 1e-64
Identities = 130/218 (59%), Positives = 150/218 (68%), Gaps = 26/218 (11%)
Frame = +1
Query: 289 DLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQGLEEFKN 348
D+P + I +TDNF + K+GEGGFGPVYKG L G EIA KRLS SGQG+ EF
Sbjct: 301 DVPLFDMLTITAATDNFLLNNKIGEGGFGPVYKGKLVGGQEIAVKRLSSLSGQGITEFIT 480
Query: 349 EVIFIAKLQHRNLVKLL--------------------------DEEKHKHLDWKLRLSII 382
EV IAKLQHRNLVKLL D+ K K LDW R +II
Sbjct: 481 EVKLIAKLQHRNLVKLLGCCIKGQEKLLVYEYVVNGSLNSFIFDQIKSKLLDWPRRFNII 660
Query: 383 KGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKRVI 442
GIARGLLYLH+DS LR+IHRDLKASNVLLD+++NPKISDFG+ARAF DQ T RV+
Sbjct: 661 LGIARGLLYLHQDSRLRIIHRDLKASNVLLDEKLNPKISDFGMARAFGGDQTEGNTNRVV 840
Query: 443 GTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRN 480
GTYGYMAPEYA G FS+KSDVFSFG+L+LEI+ G +N
Sbjct: 841 GTYGYMAPEYAFDGNFSIKSDVFSFGILLLEIVCGIKN 954
>TC222220 weakly similar to UP|Q9ZP16 (Q9ZP16) Receptor-like protein kinase,
RLK3 precursor, partial (35%)
Length = 1304
Score = 237 bits (604), Expect = 1e-62
Identities = 125/221 (56%), Positives = 148/221 (66%), Gaps = 26/221 (11%)
Frame = +3
Query: 295 LTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQGLEEFKNEVIFIA 354
L I+ +T+ FS ++GEGGFG VYKG LPDG EIA K+LS++SGQG EFKNE++ IA
Sbjct: 195 LVTIEAATNKFSYEKRIGEGGFGVVYKGVLPDGREIAVKKLSKSSGQGANEFKNEILLIA 374
Query: 355 KLQHRNLVKLL--------------------------DEEKHKHLDWKLRLSIIKGIARG 388
KLQHRNLV LL D + K L+W R II+GIA+G
Sbjct: 375 KLQHRNLVTLLGFCLEEHEKMLIYEFVSNKSLDYFLFDSHRSKQLNWSERYKIIEGIAQG 554
Query: 389 LLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKRVIGTYGYM 448
+ YLHE S L+VIHRDLK SNVLLD MNPKISDFG+AR DQ KT R++GTYGYM
Sbjct: 555 ISYLHEHSRLKVIHRDLKPSNVLLDSNMNPKISDFGMARIVAIDQLQGKTNRIVGTYGYM 734
Query: 449 APEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHN 489
+PEYAM G FS KSDVFSFGV+VLEII KRN S+H+
Sbjct: 735 SPEYAMHGQFSEKSDVFSFGVIVLEIISAKRNTRSVFSDHD 857
>TC209989 similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kinase homolog
RK20-1, partial (32%)
Length = 738
Score = 223 bits (568), Expect = 2e-58
Identities = 116/208 (55%), Positives = 144/208 (68%), Gaps = 26/208 (12%)
Frame = +3
Query: 298 IQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQGLEEFKNEVIFIAKLQ 357
I+ +T+NFS++ KLG+GGFG VYKGTL DG EIA KRLS S QG EFKNE++ +LQ
Sbjct: 114 IKFATNNFSDANKLGQGGFGIVYKGTLSDGQEIAIKRLSINSNQGETEFKNEILLTGRLQ 293
Query: 358 HRNLVKLL--------------------------DEEKHKHLDWKLRLSIIKGIARGLLY 391
HRNLV+LL D K +L+ ++R II+GIARGLLY
Sbjct: 294 HRNLVRLLGFCFARRERLLIYEFVPNKSLDYFIFDPNKRVNLN*EIRYKIIRGIARGLLY 473
Query: 392 LHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKRVIGTYGYMAPE 451
LHEDS L V+HRDLK SN+LLD E+NPKISDFG+AR FE +Q T ++GT+GYMAPE
Sbjct: 474 LHEDSRLNVVHRDLKTSNILLDGELNPKISDFGMARLFEINQTEASTTTIVGTFGYMAPE 653
Query: 452 YAMAGLFSVKSDVFSFGVLVLEIIYGKR 479
Y G FS+KSDVFSFGV++LEI+ G+R
Sbjct: 654 YIKHGQFSIKSDVFSFGVMILEIVCGQR 737
>TC221391 similar to UP|O81906 (O81906) Serine/threonine kinase-like protein
, partial (25%)
Length = 642
Score = 211 bits (536), Expect = 8e-55
Identities = 110/183 (60%), Positives = 127/183 (69%), Gaps = 26/183 (14%)
Frame = +2
Query: 327 GTEIAAKRLSETSGQGLEEFKNEVIFIAKLQHRNLVKLL--------------------- 365
G E+A KRLS S QGLEEFKNE++ IAKLQHRNLV+LL
Sbjct: 2 GEEVAVKRLSRKSSQGLEEFKNEMVLIAKLQHRNLVRLLGCCIQGEEKILVYEYLPNKSL 181
Query: 366 -----DEEKHKHLDWKLRLSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKI 420
D K LDW R II+GIARGLLYLH DS LR+IHRDLKASN+LLD+ MNPKI
Sbjct: 182 DCFLFDPVKQTQLDWAKRFEIIEGIARGLLYLHRDSRLRIIHRDLKASNILLDESMNPKI 361
Query: 421 SDFGLARAFEKDQCHTKTKRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRN 480
SDFGLAR F +Q T RV+GTYGYM+PEYAM GLFS+KSDV+SFGVL+LEI+ G++N
Sbjct: 362 SDFGLARIFGGNQNEANTNRVVGTYGYMSPEYAMEGLFSIKSDVYSFGVLLLEIMSGRKN 541
Query: 481 GDF 483
F
Sbjct: 542 TSF 550
>TC216865 similar to UP|Q9LPG0 (Q9LPG0) T3F20.24 protein, partial (28%)
Length = 1358
Score = 191 bits (486), Expect = 5e-49
Identities = 103/198 (52%), Positives = 126/198 (63%), Gaps = 27/198 (13%)
Frame = +1
Query: 310 KLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQGLEEFKNEVIFIAKLQHRNLVKLLD--- 366
K+GEGGFGPVYKG DGT IA K+LS S QG EF NE+ I+ LQH +LVKL
Sbjct: 10 KIGEGGFGPVYKGCFSDGTLIAVKQLSSKSRQGNREFLNEIGMISALQHPHLVKLYGCCV 189
Query: 367 ------------------------EEKHKHLDWKLRLSIIKGIARGLLYLHEDSPLRVIH 402
EE LDW R I GIARGL YLHE+S L+++H
Sbjct: 190 EGDQLLLVYEYMENNSLARALFGAEEHQIKLDWTTRYKICVGIARGLAYLHEESRLKIVH 369
Query: 403 RDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKRVIGTYGYMAPEYAMAGLFSVKS 462
RD+KA+NVLLD ++NPKISDFGLA+ E+D H T R+ GT+GYMAPEYAM G + K+
Sbjct: 370 RDIKATNVLLDQDLNPKISDFGLAKLDEEDNTHIST-RIAGTFGYMAPEYAMHGYLTDKA 546
Query: 463 DVFSFGVLVLEIIYGKRN 480
DV+SFG++ LEII G+ N
Sbjct: 547 DVYSFGIVALEIINGRSN 600
>TC232657 weakly similar to UP|O22580 (O22580) Receptor-like serine/threonine
kinase, partial (35%)
Length = 1208
Score = 190 bits (483), Expect = 1e-48
Identities = 99/224 (44%), Positives = 142/224 (63%), Gaps = 26/224 (11%)
Frame = +3
Query: 293 IPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQGLEEFKNEVIF 352
+P V++++T+ F+E+ KLG+GG G VYKG +PDG +A KRLS + Q E F EV
Sbjct: 174 MPYEVLEKATNYFNEANKLGQGGSGSVYKGVMPDGNTVAIKRLSYNTTQWAEHFFTEVNL 353
Query: 353 IAKLQHRNLVKLL-----------------DEEKHKH---------LDWKLRLSIIKGIA 386
I+ + H+NLVKLL ++ H H L W++R II GIA
Sbjct: 354 ISGIHHKNLVKLLGCSITGPESLLVYEYVPNQSLHDHFSVRRTSQPLTWEMRQKIILGIA 533
Query: 387 RGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKRVIGTYG 446
G+ YLHE+S +R+IHRD+K SN+LL+++ PKI+DFGLAR F +D+ H T + GT G
Sbjct: 534 EGMAYLHEESHVRIIHRDIKLSNILLEEDFTPKIADFGLARLFPEDKSHIST-AIAGTLG 710
Query: 447 YMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHNL 490
YMAPEY + G + K+DV+SFGVLV+EI+ GK+ + ++ +L
Sbjct: 711 YMAPEYIVRGKLTEKADVYSFGVLVIEIVSGKKISSYIMNSSSL 842
>TC208424 weakly similar to UP|O65466 (O65466) Serine/threonine kinase-like
protein , partial (34%)
Length = 913
Score = 181 bits (460), Expect = 5e-46
Identities = 87/125 (69%), Positives = 99/125 (78%)
Frame = +2
Query: 364 LLDEEKHKHLDWKLRLSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDF 423
L D K K LDW R II GIARG+ YLHEDS LR+IHRDLKASNVLLD+ MNPKISDF
Sbjct: 53 LFDPVKQKELDWSRRYKIIVGIARGIQYLHEDSQLRIIHRDLKASNVLLDENMNPKISDF 232
Query: 424 GLARAFEKDQCHTKTKRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDF 483
G+A+ F+ DQ T R++GTYGYM+PEYAM G FSVKSDVFSFGVLVLEI+ GK+N DF
Sbjct: 233 GMAKIFQADQTQVNTGRIVGTYGYMSPEYAMRGQFSVKSDVFSFGVLVLEIVSGKKNTDF 412
Query: 484 FLSEH 488
+ S H
Sbjct: 413 YQSNH 427
>TC221114 similar to UP|Q8RZ15 (Q8RZ15) Receptor kinase-like protein, partial
(29%)
Length = 946
Score = 177 bits (450), Expect = 7e-45
Identities = 94/190 (49%), Positives = 117/190 (61%), Gaps = 26/190 (13%)
Frame = +3
Query: 282 REDSFNGDLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQ 341
+ N ++ +I +T NFS + KLG+GGFGPVYKG LPDG EIA KRLS SGQ
Sbjct: 375 KRSKVNYEMQIFSFPIIAAATGNFSVANKLGQGGFGPVYKGVLPDGQEIAIKRLSSRSGQ 554
Query: 342 GLEEFKNEVIFIAKLQHRNLVKL--------------------------LDEEKHKHLDW 375
GL EFKNE +AKLQH NLV+L D ++ + + W
Sbjct: 555 GLVEFKNEAELVAKLQHTNLVRLSGLCIQNEENILIYEYLPNKSLDFHLFDSKRREKIVW 734
Query: 376 KLRLSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCH 435
+ R +II+GIA GL+YLH S L+VIHRDLKA N+LLD EMNPKISDFG+A + +
Sbjct: 735 EKRFNIIEGIAHGLIYLHHFSRLKVIHRDLKAGNILLDYEMNPKISDFGMAVILDSEVVE 914
Query: 436 TKTKRVIGTY 445
KTKRV+GTY
Sbjct: 915 VKTKRVVGTY 944
>TC222544 weakly similar to UP|O23743 (O23743) SFR1 protein precursor ,
partial (27%)
Length = 861
Score = 174 bits (441), Expect = 8e-44
Identities = 94/169 (55%), Positives = 113/169 (66%), Gaps = 27/169 (15%)
Frame = +3
Query: 349 EVIFIAKLQHRNLVKLL--------------------------DEEKHKHLDWKLRLSII 382
EV+ I+KLQHRNLV+LL D + K LDWK R +II
Sbjct: 3 EVVVISKLQHRNLVRLLGCCIERDEQMLVYEFMPNKSLDSFLFDPLQRKILDWKKRFNII 182
Query: 383 KGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEK-DQCHTKTKRV 441
+GIARG+LYLH DS LR+IHRDLKASN+LLDDEM+PKISDFGLAR D TKRV
Sbjct: 183 EGIARGILYLHRDSRLRIIHRDLKASNILLDDEMHPKISDFGLARIVRSGDDDEANTKRV 362
Query: 442 IGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHNL 490
+GTYGYM PEYAM G+FS KSDV+ VL+LEI+ G+RN F+ +E +L
Sbjct: 363 VGTYGYMPPEYAMEGIFSEKSDVYXXXVLLLEIVSGRRNTSFYNNEQSL 509
>TC221511 similar to UP|Q40096 (Q40096) Receptor protein kinase, partial
(21%)
Length = 765
Score = 170 bits (430), Expect = 2e-42
Identities = 83/117 (70%), Positives = 96/117 (81%)
Frame = +3
Query: 373 LDWKLRLSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKD 432
LDW R +II GIA+GLLYLH+DS LR+IHRDLKASNVLLD E+NPKISDFG+AR F D
Sbjct: 3 LDWSKRFNIICGIAKGLLYLHQDSRLRIIHRDLKASNVLLDSELNPKISDFGMARIFGVD 182
Query: 433 QCHTKTKRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHN 489
Q TKR++GTYGYMAPEYA GLFSVKSDVFSFGVL+LEII GKR+ ++ H+
Sbjct: 183 QQEGNTKRIVGTYGYMAPEYATDGLFSVKSDVFSFGVLLLEIISGKRSRGYYNQNHS 353
>TC230095 weakly similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kinase
homolog RK20-1, partial (30%)
Length = 819
Score = 168 bits (426), Expect = 4e-42
Identities = 89/162 (54%), Positives = 107/162 (65%), Gaps = 26/162 (16%)
Frame = +3
Query: 351 IFIAKLQHRNLVKLL--------------------------DEEKHKHLDWKLRLSIIKG 384
+ IAKLQH+NLV+L+ D +KH+ L W R I+KG
Sbjct: 6 LLIAKLQHKNLVRLVGFCQEDREKILIYEYVPNKSLDHFLFDSQKHRQLTWSERFKIVKG 185
Query: 385 IARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKRVIGT 444
IARG+LYLHEDS L++IHRD+K SNVLLD +NPKISDFG+AR DQ T RV+GT
Sbjct: 186 IARGILYLHEDSRLKIIHRDIKPSNVLLDYGINPKISDFGMARMVATDQIQGCTNRVVGT 365
Query: 445 YGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLS 486
YGYM+PEYAM G FS KSDVFSFGV+VL+II GK+N F S
Sbjct: 366 YGYMSPEYAMHGQFSEKSDVFSFGVMVLDIISGKKNSCSFES 491
>TC208545 weakly similar to UP|Q9S972 (Q9S972) ARK2 product/receptor-like
serine/threonine protein kinase ARK2, partial (20%)
Length = 1253
Score = 168 bits (425), Expect = 6e-42
Identities = 79/119 (66%), Positives = 99/119 (82%)
Frame = +2
Query: 366 DEEKHKHLDWKLRLSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGL 425
D + K LDWK R +II+GI++GLLYLH+ S L+VIHRDLKASN+LLD+ MNPKISDFGL
Sbjct: 326 DGTRSKLLDWKKRFNIIEGISQGLLYLHKYSRLKVIHRDLKASNILLDENMNPKISDFGL 505
Query: 426 ARAFEKDQCHTKTKRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFF 484
AR F + + T T R++GTYGYM+PEYAM G+FSVKSDV+SFGVL+LEI+ G+RN F+
Sbjct: 506 ARMFTRQESTTNTSRIVGTYGYMSPEYAMEGVFSVKSDVYSFGVLLLEIVSGRRNTSFY 682
>TC213718 similar to UP|Q70I29 (Q70I29) S-receptor kinase-like protein 2,
partial (20%)
Length = 525
Score = 167 bits (422), Expect = 1e-41
Identities = 82/116 (70%), Positives = 95/116 (81%)
Frame = +1
Query: 378 RLSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTK 437
RL II GIARGLLYLH+DS LR+IHRDLK SN+LLD++MNPKISDFGLAR F DQ
Sbjct: 10 RLQIIDGIARGLLYLHQDSRLRIIHRDLKVSNILLDNDMNPKISDFGLARTFGGDQAEAN 189
Query: 438 TKRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHNLETL 493
T RV+GTYGYM PEYA+ G FS+KSDVFSFGV+VLEII G++N +F SEH+L L
Sbjct: 190 TNRVMGTYGYMPPEYALHGRFSIKSDVFSFGVIVLEIISGRKNRNFQDSEHHLNLL 357
>TC232399 similar to UP|Q39086 (Q39086) Receptor kinase , partial (23%)
Length = 1167
Score = 167 bits (422), Expect = 1e-41
Identities = 83/132 (62%), Positives = 99/132 (74%)
Frame = +2
Query: 362 VKLLDEEKHKHLDWKLRLSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKIS 421
V LD K K LDW+ R II+GI RGLLYLH DS L++IHRDLKASNVLL + +NPKIS
Sbjct: 314 VYFLDPSKSKLLDWRKRCGIIEGIGRGLLYLHRDSRLKIIHRDLKASNVLLYEALNPKIS 493
Query: 422 DFGLARAFEKDQCHTKTKRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNG 481
DFG+AR F + T RV+GTYGYM+PEYAM GLFS KSDVFSFGVLV+EI+ G+RN
Sbjct: 494 DFGMARIFGGTEDQANTNRVVGTYGYMSPEYAMQGLFSEKSDVFSFGVLVIEIVSGRRNS 673
Query: 482 DFFLSEHNLETL 493
F+ ++ L L
Sbjct: 674 RFYDDDNALSLL 709
>TC227811 similar to PIR|JQ1674|JQ1674 protein kinase TMK1 , receptor type
precursor - Arabidopsis thaliana {Arabidopsis thaliana;}
, partial (40%)
Length = 1436
Score = 166 bits (419), Expect = 3e-41
Identities = 92/217 (42%), Positives = 130/217 (59%), Gaps = 30/217 (13%)
Frame = +2
Query: 293 IPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLS--ETSGQGLEEFKNEV 350
I + V++ TDNFSE LG+GGFG VY+G L DGT IA KR+ +G+G EFK+E+
Sbjct: 74 ISIQVLKNVTDNFSEKNVLGQGGFGTVYRGELHDGTRIAVKRMECGAIAGKGAAEFKSEI 253
Query: 351 IFIAKLQHRNLVKLLD----------------------------EEKHKHLDWKLRLSII 382
+ K++HR+LV LL EE + L+W RL+I
Sbjct: 254 AVLTKVRHRHLVSLLGYCLDGNEKLLVYEYMPQGTLSRHLFDWPEEGLEPLEWNRRLTIA 433
Query: 383 KGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKRVI 442
+ARG+ YLH + IHRDLK SN+LL D+M K++DFGL R + + +T R+
Sbjct: 434 LDVARGVEYLHGLAHQSFIHRDLKPSNILLGDDMRAKVADFGLVRLAPEGKASIET-RIA 610
Query: 443 GTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKR 479
GT+GY+APEYA+ G + K DVFSFGV+++E+I G++
Sbjct: 611 GTFGYLAPEYAVTGRVTTKVDVFSFGVILMELITGRK 721
>TC228113 similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kinase homolog
RK20-1, partial (53%)
Length = 1492
Score = 165 bits (418), Expect = 4e-41
Identities = 83/128 (64%), Positives = 98/128 (75%)
Frame = +1
Query: 366 DEEKHKHLDWKLRLSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGL 425
D K L+W++R II G+ARGLLYLHEDS LR+IHRDLKASN+LL++EMNPKI+DFG+
Sbjct: 523 DPTKKAQLNWEMRYKIITGVARGLLYLHEDSHLRIIHRDLKASNILLNEEMNPKIADFGM 702
Query: 426 ARAFEKDQCHTKTKRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFL 485
AR DQ T R++GTYGYMAPEYAM G FS+KSDVFSFGVLVLEII G +N
Sbjct: 703 ARLVLMDQTQANTNRIVGTYGYMAPEYAMHGQFSMKSDVFSFGVLVLEIISGHKNSGIRH 882
Query: 486 SEHNLETL 493
E N+E L
Sbjct: 883 GE-NVEDL 903
Score = 89.7 bits (221), Expect = 3e-18
Identities = 44/68 (64%), Positives = 54/68 (78%)
Frame = +1
Query: 298 IQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQGLEEFKNEVIFIAKLQ 357
I+ +T++FS+S KLG+GGFG VY+G L DG IA KRLS S QG EFKNEV+ +AKLQ
Sbjct: 133 IRVATEDFSDSNKLGQGGFGAVYRGRLSDGQMIAVKRLSRESSQGDTEFKNEVLLVAKLQ 312
Query: 358 HRNLVKLL 365
HRNLV+LL
Sbjct: 313 HRNLVRLL 336
>AW203661
Length = 403
Score = 162 bits (411), Expect = 2e-40
Identities = 76/119 (63%), Positives = 94/119 (78%)
Frame = +1
Query: 366 DEEKHKHLDWKLRLSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGL 425
D K + LDW R+ II GIA+G+LYLH+ S R+IHRDLKASN+LLD MNPKISDFG+
Sbjct: 1 DATKRRMLDWGTRVRIIDGIAQGILYLHQYSRFRIIHRDLKASNILLDTNMNPKISDFGM 180
Query: 426 ARAFEKDQCHTKTKRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFF 484
AR F ++ T R++GTYGYM+PEYAM GLFS+KSDVFSFGVL+LEI+ GK+N F+
Sbjct: 181 ARIFGDNELQANTNRIVGTYGYMSPEYAMEGLFSIKSDVFSFGVLLLEILSGKKNTGFY 357
>TC230548 weakly similar to UP|Q8LLI4 (Q8LLI4) S-locus receptor-like kinase
RLK14, partial (20%)
Length = 692
Score = 161 bits (407), Expect = 7e-40
Identities = 79/107 (73%), Positives = 89/107 (82%)
Frame = +3
Query: 378 RLSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTK 437
R II GIARGLLYLH+DS LR+IHRDLKASNVLLD+EMNPKISDFGLAR DQ +
Sbjct: 3 RFGIINGIARGLLYLHQDSRLRIIHRDLKASNVLLDNEMNPKISDFGLARMCGGDQIEGE 182
Query: 438 TKRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFF 484
T RV+GTYGYMAPEYA G+FS+KSDVFSFGVL+LEI+ GK+N F
Sbjct: 183 TSRVVGTYGYMAPEYAFDGIFSIKSDVFSFGVLLLEIVSGKKNSRLF 323
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.136 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,295,857
Number of Sequences: 63676
Number of extensions: 374771
Number of successful extensions: 3407
Number of sequences better than 10.0: 917
Number of HSP's better than 10.0 without gapping: 2826
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2929
length of query: 493
length of database: 12,639,632
effective HSP length: 101
effective length of query: 392
effective length of database: 6,208,356
effective search space: 2433675552
effective search space used: 2433675552
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC147431.1


