

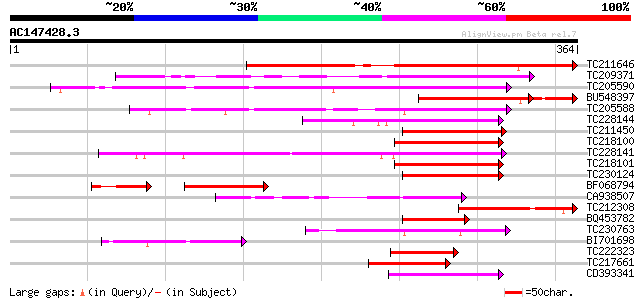
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147428.3 - phase: 0
(364 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC211646 homologue to GB|AAP37701.1|30725358|BT008342 At4g32890 ... 285 2e-77
TC209371 similar to PIR|T52104|T52104 GATA-binding transcription... 170 1e-42
TC205590 similar to UP|Q76DY0 (Q76DY0) AG-motif binding protein-... 168 4e-42
BU548397 138 2e-41
TC205588 similar to UP|Q9FH57 (Q9FH57) GATA-binding transcriptio... 143 1e-34
TC228144 homologue to UP|Q76DY3 (Q76DY3) AG-motif binding protei... 122 3e-28
TC211450 similar to UP|Q76DY3 (Q76DY3) AG-motif binding protein-... 120 1e-27
TC218100 similar to UP|Q9AVU3 (Q9AVU3) GATA-1 zinc finger protei... 119 2e-27
TC228141 similar to GB|AAL77730.1|18700240|AY078029 AT3g54810/F2... 119 2e-27
TC218101 similar to GB|AAL77730.1|18700240|AY078029 AT3g54810/F2... 119 2e-27
TC230124 similar to UP|Q76DY1 (Q76DY1) AG-motif binding protein-... 117 1e-26
BF068794 77 8e-21
CA938507 homologue to PIR|T52104|T521 GATA-binding transcription... 95 4e-20
TC212308 similar to UP|Q76DY1 (Q76DY1) AG-motif binding protein-... 93 2e-19
BQ453782 77 1e-14
TC230763 62 5e-10
BI701698 similar to GP|20197025|gb| Expressed protein {Arabidops... 61 9e-10
TC222323 similar to UP|Q9FJ10 (Q9FJ10) Gb|AAF63824.1, partial (39%) 60 2e-09
TC217661 similar to UP|Q9FJ10 (Q9FJ10) Gb|AAF63824.1, partial (47%) 59 3e-09
CD393341 59 3e-09
>TC211646 homologue to GB|AAP37701.1|30725358|BT008342 At4g32890 {Arabidopsis
thaliana;} , partial (26%)
Length = 778
Score = 285 bits (730), Expect = 2e-77
Identities = 147/216 (68%), Positives = 168/216 (77%), Gaps = 4/216 (1%)
Frame = +2
Query: 153 FSSEDLHKLQLISGLKAPNNVASKPYEESNPTVHS-QVSVPAKARSKRSRVPPCNWTSRL 211
FSSEDL KLQLI+G++A N+ AS + NP + + QVSV KARSKR+R PPCNWTSRL
Sbjct: 2 FSSEDLQKLQLITGVRAQNDAASSETRDPNPVMFNPQVSVRGKARSKRTRGPPCNWTSRL 181
Query: 212 LVLSPTTTTTTTTTTSSHSDTMAPPKKPSPRKRDPNDGGEGRKCLHCATDKTPQWRTGPL 271
+VLSP T +++ SSHS G EGRKCLHCATDKTPQWRTGP+
Sbjct: 182 VVLSPNTKSSS----SSHSGA--------------EGGSEGRKCLHCATDKTPQWRTGPM 307
Query: 272 GPKTLCNACGVRYKSGRLVPEYRPAASPTFVLTKHSNSHRKVQELRRQKEMMRA--QQHQ 329
GPKTLCNACGVRYKSGRLVPEYRPAASPTFVLTKHSNSHRKV ELRRQKEM++ QQHQ
Sbjct: 308 GPKTLCNACGVRYKSGRLVPEYRPAASPTFVLTKHSNSHRKVLELRRQKEMVKVQHQQHQ 487
Query: 330 LLQLQHHHSIMFEGP-SNGDDYLIHQHVGPDFTHLI 364
LQLQH ++MF+ P SNG+D+LIHQHVGP+FTHLI
Sbjct: 488 FLQLQHQQNMMFDVPSSNGEDFLIHQHVGPNFTHLI 595
>TC209371 similar to PIR|T52104|T52104 GATA-binding transcription factor
homolog 2 [imported] - Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (40%)
Length = 857
Score = 170 bits (430), Expect = 1e-42
Identities = 108/270 (40%), Positives = 139/270 (51%), Gaps = 1/270 (0%)
Frame = +2
Query: 69 SDHFIVEDLFDFSNEDVAIEDPTFEESPPTNSNDSPPLETNPTSNFFTDNSCQNSADGPF 128
SD ++D DFSN D PP NS P + +P NFF +
Sbjct: 29 SDCLHIDDFLDFSNITTTTTDTHHHFPPPQNS---PSISHDP--NFFLNFP--------- 166
Query: 129 SGELSVPYDDLAELEWVSKFAEESFSSEDLHKLQLISGLKAPNNVASKPYEESNPTVHSQ 188
SVP D+ ELEW+S+F + +S + P ++ S H+
Sbjct: 167 ----SVPSDEAVELEWLSQFVNDEATS--------FHNIPPPASIGS----------HTT 280
Query: 189 VSVPAKARSKRSRVPPCNWTSRLLVLSPTTTTTTTTTTSSHSDTMAPPKKPSPRKRDPND 248
+ R+ + P +SS S +A + +R+ +
Sbjct: 281 PFLSNNNRNDNNNEYP--------------------KSSSSSPVLAGKSRA---RREGSV 391
Query: 249 GGEG-RKCLHCATDKTPQWRTGPLGPKTLCNACGVRYKSGRLVPEYRPAASPTFVLTKHS 307
G+G R+C HCATDKTPQWRTGPLGPKTLCNACGVR+KSGRLVPEYRPAASPTFV+T+HS
Sbjct: 392 TGDGVRRCSHCATDKTPQWRTGPLGPKTLCNACGVRFKSGRLVPEYRPAASPTFVMTQHS 571
Query: 308 NSHRKVQELRRQKEMMRAQQHQLLQLQHHH 337
NSHRKV ELRRQKE++R QQ + HH
Sbjct: 572 NSHRKVMELRRQKELLRHQQQEQCYRHTHH 661
>TC205590 similar to UP|Q76DY0 (Q76DY0) AG-motif binding protein-4, partial
(25%)
Length = 1249
Score = 168 bits (425), Expect = 4e-42
Identities = 117/307 (38%), Positives = 159/307 (51%), Gaps = 11/307 (3%)
Frame = +3
Query: 27 FIIFF-------ASTSKPNNNNN-LTQPMEAQEFFQSDNNSNTNNVNTAASDHFIVEDLF 78
F+ FF A KP+ + Q M ++ F N+NT AA + F V+DLF
Sbjct: 90 FLFFFTEMEVAVAKALKPSLRREFIVQQMLCEDIFSL--NANT----VAAGEDFSVDDLF 251
Query: 79 DFSNEDVAIEDPT-FEESPPTNSNDSPPLETNPTSNFFTDNSCQNSADGPFSGELSVPYD 137
DFSN + E ++E + S E + SN S S D FS EL+VP
Sbjct: 252 DFSNGSLHNEQQQEYDEGKQSLSASEDRGEDDCNSN-----STGVSYDSLFSTELAVPAG 416
Query: 138 DLAELEWVSKFAEESFSSEDLHKLQLISGLKAPNNVASKPYEESNPTVHSQVSVPAKARS 197
DL +LEWVS F ++S +L L + +A V +P + P ++ + KAR+
Sbjct: 417 DLEDLEWVSHFVDDSLP--ELSLLYPVRSEEANRFVEPEPSAKKTPCFPWEMKITTKART 590
Query: 198 KRSRVPPCN--WTSRLLVLSPTTTTTTTTTTSSHSDTMAPPKKPSPRKRDPNDGGEGRKC 255
R+R P + W+ VLS ++ ++ ++ SS K+ + P R+C
Sbjct: 591 VRNRKPSNSRMWSVG*PVLSLPSSPSSPSSCSSSVREPPAKKQKKQAQAQPVGAQIQRRC 770
Query: 256 LHCATDKTPQWRTGPLGPKTLCNACGVRYKSGRLVPEYRPAASPTFVLTKHSNSHRKVQE 315
HC KTPQWRTGPLG KT NACGVRYKSGRL EYRPA SPTF HSNSHRKV E
Sbjct: 771 SHCHVQKTPQWRTGPLGAKTXXNACGVRYKSGRLFSEYRPACSPTFCSDIHSNSHRKVLE 950
Query: 316 LRRQKEM 322
+R++KE+
Sbjct: 951 IRKRKEV 971
>BU548397
Length = 605
Score = 138 bits (347), Expect(2) = 2e-41
Identities = 66/78 (84%), Positives = 69/78 (87%), Gaps = 4/78 (5%)
Frame = -2
Query: 263 TPQWRTGPLGPKTLCNACGVRYKSGRLVPEYRPAASPTFVLTKHSNSHRKVQELRRQKEM 322
TPQWRTGP+GPKT CNACGVRYKSGRLVPEYRPAASPTFVLTKHSNSHRKV ELRRQKEM
Sbjct: 604 TPQWRTGPMGPKTXCNACGVRYKSGRLVPEYRPAASPTFVLTKHSNSHRKVXELRRQKEM 425
Query: 323 MRAQ----QHQLLQLQHH 336
+R+Q QHQ LQHH
Sbjct: 424 VRSQQHHHQHQQQFLQHH 371
Score = 49.3 bits (116), Expect(2) = 2e-41
Identities = 22/31 (70%), Positives = 26/31 (82%)
Frame = -3
Query: 334 QHHHSIMFEGPSNGDDYLIHQHVGPDFTHLI 364
QHH ++MF+ SNGDDYLIHQ+VGPDF LI
Sbjct: 348 QHHQNMMFD-VSNGDDYLIHQYVGPDFRQLI 259
>TC205588 similar to UP|Q9FH57 (Q9FH57) GATA-binding transcription
factor-like protein (At5g66320), partial (25%)
Length = 1084
Score = 143 bits (360), Expect = 1e-34
Identities = 101/274 (36%), Positives = 132/274 (47%), Gaps = 29/274 (10%)
Frame = +3
Query: 78 FDFSNEDVAIE--DPTFEESPPTNSNDSPPLETNPTSNFFTDNSCQNSADGPFSGELSVP 135
FDFSN + E EE +++ SN NS S D FS EL+VP
Sbjct: 3 FDFSNGSLHNEHQQECDEEKQSLSASSQSQDRGEDDSN---SNSTGVSYDSLFSTELAVP 173
Query: 136 YD-----------------------DLAELEWVSKFAEESFSSEDLHKLQLISGLKAPNN 172
DL +LEWVS F ++S +L L + +A
Sbjct: 174 VTLSPFVPISISNFVF**LKFV*AGDLEDLEWVSHFVDDSLP--ELSLLYPVRSEEANRF 347
Query: 173 VASKPYEESNPTVHSQVSVPAKARSKRSRVPPCNWTSRLLVLSPTTTTTTTTTTSSHSDT 232
V +P + P ++ + +KARS R+R P +R+ L T + ++
Sbjct: 348 VEPEPSVKKTPRFPWEMKITSKARSVRNRKP----NTRVWSLGSTLLSLPSS-------- 491
Query: 233 MAPPKKPSPRKRDPNDGGEG----RKCLHCATDKTPQWRTGPLGPKTLCNACGVRYKSGR 288
PP K ++ + G R+C HC KTPQWRTGPLG KTLCNACGVRYKSGR
Sbjct: 492 --PPTKKQKKRAEAQVQPVGVQIQRRCSHCQVQKTPQWRTGPLGAKTLCNACGVRYKSGR 665
Query: 289 LVPEYRPAASPTFVLTKHSNSHRKVQELRRQKEM 322
L EYRPA SPTF HSNSHRKV E+R++KE+
Sbjct: 666 LFSEYRPACSPTFCSDIHSNSHRKVLEIRKRKEV 767
>TC228144 homologue to UP|Q76DY3 (Q76DY3) AG-motif binding protein-1, partial
(27%)
Length = 1018
Score = 122 bits (305), Expect = 3e-28
Identities = 71/161 (44%), Positives = 88/161 (54%), Gaps = 32/161 (19%)
Frame = +1
Query: 189 VSVPA-KARSKRSRVPPCNWTSRLLVLSPTTT--------TTTTTTTSSHSDTMAP---- 235
+ VP +ARSKR R N + ++SP ++ ++ SS S+ A
Sbjct: 19 IPVPCGRARSKRPRPATFNPRPAMNLISPASSFVGENMQPNVISSKASSDSENFAESQLV 198
Query: 236 ----------PKKPS---------PRKRDPNDGGEGRKCLHCATDKTPQWRTGPLGPKTL 276
PKK + P + N RKC+HC KTPQWR GP+GPKTL
Sbjct: 199 PIFLKLASGEPKKKNKVKVPLPVAPADNNQNASQPVRKCMHCEITKTPQWRAGPMGPKTL 378
Query: 277 CNACGVRYKSGRLVPEYRPAASPTFVLTKHSNSHRKVQELR 317
CNACGVRYKSGRL PEYRPAASPTF + HSNSH+KV E+R
Sbjct: 379 CNACGVRYKSGRLFPEYRPAASPTFCPSVHSNSHKKVLEMR 501
>TC211450 similar to UP|Q76DY3 (Q76DY3) AG-motif binding protein-1, partial
(23%)
Length = 825
Score = 120 bits (300), Expect = 1e-27
Identities = 52/67 (77%), Positives = 56/67 (82%)
Frame = +2
Query: 253 RKCLHCATDKTPQWRTGPLGPKTLCNACGVRYKSGRLVPEYRPAASPTFVLTKHSNSHRK 312
RKCLHC KTPQWR GP+GPKTLCNACGVRYKSGRL PEYRPAASPTF HSNSH+K
Sbjct: 182 RKCLHCEITKTPQWRAGPMGPKTLCNACGVRYKSGRLFPEYRPAASPTFCAAMHSNSHKK 361
Query: 313 VQELRRQ 319
V E+R +
Sbjct: 362 VLEMRNK 382
>TC218100 similar to UP|Q9AVU3 (Q9AVU3) GATA-1 zinc finger protein, partial
(27%)
Length = 1284
Score = 119 bits (299), Expect = 2e-27
Identities = 51/70 (72%), Positives = 58/70 (82%)
Frame = +2
Query: 248 DGGEGRKCLHCATDKTPQWRTGPLGPKTLCNACGVRYKSGRLVPEYRPAASPTFVLTKHS 307
+ G RKC+HC KTPQWR GP+GPKTLCNACGVRY+SGRL PEYRPAASPTFV + HS
Sbjct: 878 ESGSPRKCMHCEVTKTPQWREGPVGPKTLCNACGVRYRSGRLFPEYRPAASPTFVASLHS 1057
Query: 308 NSHRKVQELR 317
N H+KV E+R
Sbjct: 1058NCHKKVVEMR 1087
>TC228141 similar to GB|AAL77730.1|18700240|AY078029 AT3g54810/F28P10_210
{Arabidopsis thaliana;} , partial (23%)
Length = 1143
Score = 119 bits (299), Expect = 2e-27
Identities = 93/292 (31%), Positives = 131/292 (44%), Gaps = 30/292 (10%)
Frame = +3
Query: 58 NSNTNNVNTAASDHFIVEDLFDF--SNEDV---AIEDPTFEESPPTNSNDSPPLETNP-- 110
++N N ++ + D + +L D EDV A+E+ ++ + PPL P
Sbjct: 84 DNNFNGLSDESLDDVMDMELLDLPLDFEDVETDAVEEQDWDAQLKLLEDPPPPLGVFPLQ 263
Query: 111 -TSNFFTDNSCQNSADGPFSGELSVPYDDLAELEWVSKFAEESFSSEDLHKLQLISGLKA 169
+S F +N+ G S S+ + S +DL + Q S +
Sbjct: 264 QSSAFCGQTRNENAKLGSKSFSASLAKTVRPAYGKTIPVQKVSLKGKDLLQFQTNSPVSV 443
Query: 170 PNNVASKPYEESNPTVHSQVSVPAKARSKRSRVPPCNWTSRL-LVLSPTTTTTTTTTTSS 228
+ +S P E N V + R+KR R+ + + +L+ S
Sbjct: 444 FESSSSSPSVE-NSNFELPVIPTKRPRTKRRRLSNISLLYSIPFILTSPAFQKFQRMDFS 620
Query: 229 HSDTMAPPK-------KPSPRKRD--------------PNDGGEGRKCLHCATDKTPQWR 267
SD P K RK+D + RKCLHC KTPQWR
Sbjct: 621 KSDIQTQPSGELLCKFKKKQRKKDIPLPTNKIEMKRSSSQESVAPRKCLHCEVTKTPQWR 800
Query: 268 TGPLGPKTLCNACGVRYKSGRLVPEYRPAASPTFVLTKHSNSHRKVQELRRQ 319
GP+GPKTLCNACGVRY+SGRL EYRPA+SPTFV + HSNSH+KV E+R +
Sbjct: 801 EGPMGPKTLCNACGVRYRSGRLFAEYRPASSPTFVASLHSNSHKKVLEIRNR 956
>TC218101 similar to GB|AAL77730.1|18700240|AY078029 AT3g54810/F28P10_210
{Arabidopsis thaliana;} , partial (23%)
Length = 638
Score = 119 bits (299), Expect = 2e-27
Identities = 51/70 (72%), Positives = 58/70 (82%)
Frame = +1
Query: 248 DGGEGRKCLHCATDKTPQWRTGPLGPKTLCNACGVRYKSGRLVPEYRPAASPTFVLTKHS 307
+ G RKC+HC KTPQWR GP+GPKTLCNACGVRY+SGRL PEYRPAASPTFV + HS
Sbjct: 196 ESGPPRKCMHCEVTKTPQWREGPMGPKTLCNACGVRYRSGRLFPEYRPAASPTFVASLHS 375
Query: 308 NSHRKVQELR 317
N H+KV E+R
Sbjct: 376 NCHKKVVEMR 405
>TC230124 similar to UP|Q76DY1 (Q76DY1) AG-motif binding protein-3, partial
(26%)
Length = 894
Score = 117 bits (292), Expect = 1e-26
Identities = 50/65 (76%), Positives = 55/65 (83%)
Frame = +1
Query: 253 RKCLHCATDKTPQWRTGPLGPKTLCNACGVRYKSGRLVPEYRPAASPTFVLTKHSNSHRK 312
R+C HC +TPQWR GPLGPKTLCNACGVRYKSGRL+PEYRPA SPTFV HSNSH+K
Sbjct: 190 RRCTHCLAQRTPQWRAGPLGPKTLCNACGVRYKSGRLLPEYRPAKSPTFVSYLHSNSHKK 369
Query: 313 VQELR 317
V E+R
Sbjct: 370 VMEMR 384
>BF068794
Length = 383
Score = 76.6 bits (187), Expect(2) = 8e-21
Identities = 36/54 (66%), Positives = 42/54 (77%)
Frame = +1
Query: 113 NFFTDNSCQNSADGPFSGELSVPYDDLAELEWVSKFAEESFSSEDLHKLQLISG 166
N D +N +DG FS +L VPYDD+AELEW+S F EESFSSEDLHK+QLISG
Sbjct: 220 NTVPDIGSRNLSDGHFSDDLCVPYDDIAELEWLSNFVEESFSSEDLHKMQLISG 381
Score = 41.6 bits (96), Expect(2) = 8e-21
Identities = 22/39 (56%), Positives = 24/39 (61%)
Frame = +3
Query: 53 FQSDNNSNTNNVNTAASDHFIVEDLFDFSNEDVAIEDPT 91
F DNN+ SDHFIVEDL DFSN+DV I D T
Sbjct: 45 FTFDNNN---------SDHFIVEDLLDFSNDDVVITDAT 134
>CA938507 homologue to PIR|T52104|T521 GATA-binding transcription factor
homolog 2 [imported] - Arabidopsis thaliana, partial
(23%)
Length = 449
Score = 95.1 bits (235), Expect = 4e-20
Identities = 60/161 (37%), Positives = 79/161 (48%)
Frame = +1
Query: 133 SVPYDDLAELEWVSKFAEESFSSEDLHKLQLISGLKAPNNVASKPYEESNPTVHSQVSVP 192
SVP D+ AELEW+S+F ++ +S H + + + S + +N + P
Sbjct: 67 SVPTDEAAELEWLSQFVDDDATS--FHSFPATASIGSH----STSFLSNNNNRNDNNEYP 228
Query: 193 AKARSKRSRVPPCNWTSRLLVLSPTTTTTTTTTTSSHSDTMAPPKKPSPRKRDPNDGGEG 252
+S S PC S +A + DGG
Sbjct: 229 ---KSSLSSNIPC------------------------SSAVAGKSRARREGSVTGDGGV- 324
Query: 253 RKCLHCATDKTPQWRTGPLGPKTLCNACGVRYKSGRLVPEY 293
R+C HCA++KTPQWR GPLGPKTLCNACGVR+KSGRLVPEY
Sbjct: 325 RRCSHCASEKTPQWRAGPLGPKTLCNACGVRFKSGRLVPEY 447
>TC212308 similar to UP|Q76DY1 (Q76DY1) AG-motif binding protein-3, partial
(15%)
Length = 542
Score = 92.8 bits (229), Expect = 2e-19
Identities = 50/83 (60%), Positives = 56/83 (67%), Gaps = 7/83 (8%)
Frame = +3
Query: 289 LVPEYRPAASPTFVLTKHSNSHRKVQELRRQKEMMRAQQHQLLQLQHHHSIMFEGPSNGD 348
LVPEYRPAASPTF+ TKHSNSHRKV ELRRQKEM R Q HQ L Q S +F + GD
Sbjct: 3 LVPEYRPAASPTFMSTKHSNSHRKVLELRRQKEMQRQQHHQQLMSQ---SSIFGVSNGGD 173
Query: 349 DYLIHQ-------HVGPDFTHLI 364
++ IH H GPDF H+I
Sbjct: 174 EFSIHHHHHNHHLHCGPDFRHVI 242
>BQ453782
Length = 424
Score = 77.0 bits (188), Expect = 1e-14
Identities = 30/43 (69%), Positives = 35/43 (80%)
Frame = -1
Query: 253 RKCLHCATDKTPQWRTGPLGPKTLCNACGVRYKSGRLVPEYRP 295
R+C HC KTPQWRTGP G TLCNACG+R+K+G+L PEYRP
Sbjct: 406 RRCSHCDAIKTPQWRTGPFGRNTLCNACGIRFKAGKLYPEYRP 278
>TC230763
Length = 1058
Score = 61.6 bits (148), Expect = 5e-10
Identities = 42/141 (29%), Positives = 63/141 (43%), Gaps = 10/141 (7%)
Frame = +1
Query: 191 VPAKARSKRSRVPPCNWTSRLLVLSPTTTTTTTTTTSSHSDTMAPPKKPSPRKRDPNDGG 250
+PAK R R ++LV T T T + ++H + SP D +
Sbjct: 67 MPAKMRIMR----------KMLVSDQTDTYTNSDNNTTHKFDDQKQQLSSPLGTDNSSSN 216
Query: 251 EG--------RKCLHCATDKTPQWRTGPLGPKTLCNACGVRYKSGR--LVPEYRPAASPT 300
R C C T KTP WR+GP GPK+LCNACG+R + R + A+
Sbjct: 217 NYSNHSNNTVRVCSDCHTTKTPLWRSGPRGPKSLCNACGIRQRKARRAMAAAAASASGNG 396
Query: 301 FVLTKHSNSHRKVQELRRQKE 321
V+ + S + +L+++KE
Sbjct: 397 TVIVEAKKSVKGRNKLQKKKE 459
>BI701698 similar to GP|20197025|gb| Expressed protein {Arabidopsis
thaliana}, partial (9%)
Length = 437
Score = 60.8 bits (146), Expect = 9e-10
Identities = 38/100 (38%), Positives = 54/100 (54%), Gaps = 7/100 (7%)
Frame = +1
Query: 60 NTNNVNTAASDHFIVEDLFDFSNEDVAI-------EDPTFEESPPTNSNDSPPLETNPTS 112
N NNV A + F V+DL DFSN + + E+ EE T+ + T S
Sbjct: 121 NANNV--VAGEDFSVDDLLDFSNGEFQVGKDFDDYEEDEDEEKGSTSGSLQSQDRTEDDS 294
Query: 113 NFFTDNSCQNSADGPFSGELSVPYDDLAELEWVSKFAEES 152
N ++++ D F+GELSVP DD+A+LEWVS F ++S
Sbjct: 295 N--SNSTAGGGGDSVFAGELSVPADDVADLEWVSHFVDDS 408
>TC222323 similar to UP|Q9FJ10 (Q9FJ10) Gb|AAF63824.1, partial (39%)
Length = 368
Score = 59.7 bits (143), Expect = 2e-09
Identities = 25/44 (56%), Positives = 29/44 (65%)
Frame = +2
Query: 245 DPNDGGEGRKCLHCATDKTPQWRTGPLGPKTLCNACGVRYKSGR 288
D N + + C C T KTP WR GP GPKTLCNACG+RY+ R
Sbjct: 158 DLNVNEKKKCCADCKTTKTPLWRGGPAGPKTLCNACGIRYRKRR 289
>TC217661 similar to UP|Q9FJ10 (Q9FJ10) Gb|AAF63824.1, partial (47%)
Length = 765
Score = 59.3 bits (142), Expect = 3e-09
Identities = 24/53 (45%), Positives = 32/53 (60%)
Frame = +2
Query: 231 DTMAPPKKPSPRKRDPNDGGEGRKCLHCATDKTPQWRTGPLGPKTLCNACGVR 283
D+ + P PS ++ + + C C T KTP WR GP GPK+LCNACG+R
Sbjct: 218 DSNSNPNAPSSGNSPSSNNEQKKTCADCGTTKTPLWRGGPAGPKSLCNACGIR 376
>CD393341
Length = 612
Score = 58.9 bits (141), Expect = 3e-09
Identities = 28/74 (37%), Positives = 37/74 (49%)
Frame = -3
Query: 244 RDPNDGGEGRKCLHCATDKTPQWRTGPLGPKTLCNACGVRYKSGRLVPEYRPAASPTFVL 303
R+ N R C C T TP WRTGP GPK+LCNACG+R + R T +
Sbjct: 577 RNNNGSSTPRVCSDCNTSTTPLWRTGPKGPKSLCNACGIRQRKARRAMAEAANGLVTPIA 398
Query: 304 TKHSNSHRKVQELR 317
+ + H K ++ R
Sbjct: 397 CEKTRLHNKEKKSR 356
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.313 0.129 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,420,525
Number of Sequences: 63676
Number of extensions: 333324
Number of successful extensions: 4838
Number of sequences better than 10.0: 295
Number of HSP's better than 10.0 without gapping: 3582
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4259
length of query: 364
length of database: 12,639,632
effective HSP length: 99
effective length of query: 265
effective length of database: 6,335,708
effective search space: 1678962620
effective search space used: 1678962620
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC147428.3


