

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147364.1 - phase: 0
(200 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
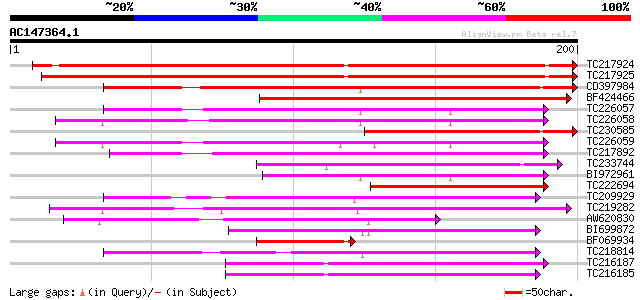
Score E
Sequences producing significant alignments: (bits) Value
TC217924 234 2e-62
TC217925 224 2e-59
CD397984 159 6e-40
BF424466 139 9e-34
TC226057 weakly similar to GB|AAG40073.2|13265596|AF324722 AT4g1... 101 3e-22
TC226058 similar to GB|AAG40073.2|13265596|AF324722 AT4g15610 {A... 100 8e-22
TC230585 92 2e-19
TC226059 similar to GB|AAG40073.2|13265596|AF324722 AT4g15610 {A... 82 2e-16
TC217892 similar to UP|Q6I7V6 (Q6I7V6) ATPase subunit 6, partial... 73 8e-14
TC233744 73 8e-14
BI972961 66 9e-12
TC222694 weakly similar to GB|AAG40073.2|13265596|AF324722 AT4g1... 65 2e-11
TC209929 weakly similar to GB|AAM70533.1|21700819|AY124824 At2g3... 61 3e-10
TC219282 weakly similar to UP|Q9FFZ7 (Q9FFZ7) Gb|AAF00668.1, par... 59 1e-09
AW620830 56 1e-08
BI699872 53 1e-07
BF069934 51 3e-07
TC218814 50 9e-07
TC216187 48 3e-06
TC216185 47 4e-06
>TC217924
Length = 843
Score = 234 bits (597), Expect = 2e-62
Identities = 119/192 (61%), Positives = 148/192 (76%)
Frame = +3
Query: 9 IDGVEGNYGRDELAMKKPPAGGGSCELVLRFLGFVLTLAAAIVVGTDKQTTIVPIKVVDS 68
++GVE E+ + KP A G C+L+LR L F +TL AAIV+ DKQT +VPI++ DS
Sbjct: 126 MEGVESK--EREVMVAKPVAVVGVCDLLLRLLAFTVTLVAAIVIAVDKQTKLVPIQLSDS 299
Query: 69 LPPLNVAVSAKWHYLSAFVYFLVANAIACTYGAISMLLTLLNRGKSKVFWGTLITIFDAL 128
PPLNV ++AKWH +SAFVYFLV NAIACTY A+S+LL L+NRGKSK W TLI + D
Sbjct: 300 FPPLNVPLTAKWHQMSAFVYFLVTNAIACTYAAMSLLLALVNRGKSKGLW-TLIAVLDTF 476
Query: 129 MVALLFSGNGAATAIGVLGYQGNSHVRWKKVCNVFDKYCHQVAASIILSQLGSLVFLLLV 188
MVALLFSGNGAA A+G+LGY+GNSHV W KVCNVF K+C Q+AASI +S +GSL FLLLV
Sbjct: 477 MVALLFSGNGAAAAVGILGYKGNSHVNWNKVCNVFGKFCDQMAASIGVSLIGSLAFLLLV 656
Query: 189 VLLPILRSRRRS 200
V +P++R RR+
Sbjct: 657 V-IPVVRLHRRT 689
>TC217925
Length = 805
Score = 224 bits (572), Expect = 2e-59
Identities = 114/189 (60%), Positives = 142/189 (74%)
Frame = +3
Query: 12 VEGNYGRDELAMKKPPAGGGSCELVLRFLGFVLTLAAAIVVGTDKQTTIVPIKVVDSLPP 71
+EG ++ M P G +L+LR L F +TL AAIV+ DKQT +VPI++ DSLPP
Sbjct: 12 MEGVESKEREVMVAKPVAVGVSDLLLRLLAFTVTLVAAIVIAVDKQTKVVPIQLSDSLPP 191
Query: 72 LNVAVSAKWHYLSAFVYFLVANAIACTYGAISMLLTLLNRGKSKVFWGTLITIFDALMVA 131
L+V ++AKWH +SA VYFLV NAIACTY +S+LL L+NRGKSK W TLI + DA MVA
Sbjct: 192 LDVPLTAKWHQMSAIVYFLVTNAIACTYAVLSLLLALVNRGKSKGLW-TLIAVLDAFMVA 368
Query: 132 LLFSGNGAATAIGVLGYQGNSHVRWKKVCNVFDKYCHQVAASIILSQLGSLVFLLLVVLL 191
LLFSGNGAA A+GVLGY+GNSHV W KVCNVF K+C Q+AASI +S +GSL FLLLV+ +
Sbjct: 369 LLFSGNGAAAAVGVLGYKGNSHVNWNKVCNVFGKFCDQMAASIGVSLIGSLAFLLLVI-I 545
Query: 192 PILRSRRRS 200
P +R RR+
Sbjct: 546 PGVRLHRRN 572
>CD397984
Length = 643
Score = 159 bits (403), Expect = 6e-40
Identities = 85/169 (50%), Positives = 115/169 (67%), Gaps = 2/169 (1%)
Frame = -3
Query: 34 ELVLRFLGFVLTLAAAIVVGTDKQTTIVPIKVVDSLPPLNVAVSAKWHYLSAFVYFLVAN 93
+L+LRFLG LTL A I+VG D +T ++ S + +AKW Y+SA V+FLV N
Sbjct: 626 DLLLRFLGLSLTLVAVIIVGVDNETKVI------SYAEMQFKATAKWEYVSAIVFFLVIN 465
Query: 94 AIACTYGAISMLLTLLNRGKSKVFWGTLI--TIFDALMVALLFSGNGAATAIGVLGYQGN 151
AIAC+Y A S+++TL+ R + TL+ T D +M+ALLFS NGAA A+GV+ +GN
Sbjct: 464 AIACSYAAASLVITLMARSHGRKNDVTLLGLTALDLVMMALLFSANGAACAVGVIAQKGN 285
Query: 152 SHVRWKKVCNVFDKYCHQVAASIILSQLGSLVFLLLVVLLPILRSRRRS 200
SHV+W KVCNVFD YC V A+++LS +GS VFLLL VLL +L+ R+
Sbjct: 284 SHVQWMKVCNVFDAYCRHVTAALVLSIIGSTVFLLL-VLLSVLKLHYRT 141
>BF424466
Length = 419
Score = 139 bits (350), Expect = 9e-34
Identities = 67/110 (60%), Positives = 85/110 (76%)
Frame = +1
Query: 89 FLVANAIACTYGAISMLLTLLNRGKSKVFWGTLITIFDALMVALLFSGNGAATAIGVLGY 148
++ ANAIAC Y +S+LLTL NR K K TLIT+ D +MVALLFSGNGAA A+G+LG
Sbjct: 4 YVGANAIACAYAILSLLLTLANRRKGKGTMETLITVLDTVMVALLFSGNGAAMAVGLLGL 183
Query: 149 QGNSHVRWKKVCNVFDKYCHQVAASIILSQLGSLVFLLLVVLLPILRSRR 198
QGNSHV W KVCN F K+C QVAAS+ +S LGS+ FLLL+++ P+LR ++
Sbjct: 184 QGNSHVHWNKVCNEFGKFCDQVAASLFISLLGSIAFLLLLIVPPVLRLKQ 333
>TC226057 weakly similar to GB|AAG40073.2|13265596|AF324722 AT4g15610
{Arabidopsis thaliana;} , partial (47%)
Length = 758
Score = 101 bits (251), Expect = 3e-22
Identities = 58/160 (36%), Positives = 92/160 (57%), Gaps = 3/160 (1%)
Frame = +1
Query: 34 ELVLRFLGFVLTLAAAIVVGTDKQTTIVPIKVVDSLPPLNVAVSAKWHYLSAFVYFLVAN 93
+++LRF+ F +L A +V+ T QT ++ L P V AK+ Y AFVYF+ A
Sbjct: 220 DVILRFVLFAASLVAVVVIVTGNQTEVI-------LVPQPVPWPAKFRYTPAFVYFVAAL 378
Query: 94 AIACTYGAISMLLTLLNRGKSKVFWGTLI--TIFDALMVALLFSGNGAATAIGVLGYQGN 151
++ Y I+ L +L K + L+ ++DAL++ ++ S G A + LG +GN
Sbjct: 379 SVTGLYSIITTLASLFASNKPALKTKLLLYFILWDALILGIIASATGTAGGVAYLGLKGN 558
Query: 152 SHV-RWKKVCNVFDKYCHQVAASIILSQLGSLVFLLLVVL 190
SHV W K+C+V+DK+C V ASI ++ GS+V +LL+ L
Sbjct: 559 SHVVGWNKICHVYDKFCRHVGASIAVALFGSIVTVLLIWL 678
>TC226058 similar to GB|AAG40073.2|13265596|AF324722 AT4g15610 {Arabidopsis
thaliana;} , partial (47%)
Length = 1014
Score = 99.8 bits (247), Expect = 8e-22
Identities = 62/181 (34%), Positives = 96/181 (52%), Gaps = 7/181 (3%)
Frame = +1
Query: 17 GRDELAMKKPPAGGG----SCELVLRFLGFVLTLAAAIVVGTDKQTTIVPIKVVDSLPPL 72
G E PA G +++LRFL F +L A +V+ T QT ++ + P
Sbjct: 187 GDPEYRTSSTPAPAGVDYFKFDVILRFLLFAASLVAVVVIVTANQTEVIRV-------PQ 345
Query: 73 NVAVSAKWHYLSAFVYFLVANAIACTYGAISMLLTLLNRGKSKVFWGTLI--TIFDALMV 130
V AK+ Y AFVYF+ A ++ Y I+ L +L K + L+ ++DAL++
Sbjct: 346 PVPWPAKFRYSPAFVYFVAALSVTGLYSIITTLASLFASNKPALKTKLLLYFILWDALIL 525
Query: 131 ALLFSGNGAATAIGVLGYQGNSHV-RWKKVCNVFDKYCHQVAASIILSQLGSLVFLLLVV 189
++ S G A + LG +GN HV W K+C+V+DK+C V ASI ++ GS+V +LL+
Sbjct: 526 GIIASATGTAGGVAYLGLKGNRHVVGWNKICHVYDKFCRHVGASIAVALFGSVVTVLLIW 705
Query: 190 L 190
L
Sbjct: 706 L 708
>TC230585
Length = 403
Score = 92.0 bits (227), Expect = 2e-19
Identities = 45/75 (60%), Positives = 57/75 (76%)
Frame = +2
Query: 126 DALMVALLFSGNGAATAIGVLGYQGNSHVRWKKVCNVFDKYCHQVAASIILSQLGSLVFL 185
D +M+ALLFS NGAA A+GV+ +GNSHV+W KVCNVFD YC V A+++LS GS VFL
Sbjct: 41 DLVMMALLFSANGAACAVGVIAQKGNSHVQWMKVCNVFDAYCRHVTAALVLSLFGSTVFL 220
Query: 186 LLVVLLPILRSRRRS 200
LL VLL +L+ R+
Sbjct: 221 LL-VLLSVLKLHYRT 262
>TC226059 similar to GB|AAG40073.2|13265596|AF324722 AT4g15610 {Arabidopsis
thaliana;} , partial (38%)
Length = 943
Score = 82.0 bits (201), Expect = 2e-16
Identities = 62/225 (27%), Positives = 95/225 (41%), Gaps = 51/225 (22%)
Frame = +3
Query: 17 GRDELAMKKPPAGGG----SCELVLRFLGFVLTLAAAIVVGTDKQTTIVPIKVVDSLPPL 72
G E PA G +++LRF+ F +L A +V+ T QT ++ L P
Sbjct: 39 GDPEYRTSSTPAPAGVDYFKFDVILRFVLFAASLVAVVVIVTGNQTEVI-------LVPQ 197
Query: 73 NVAVSAKWHYLSAFVYFLVANAIACTYGAISMLLTLLNRGKSK---------VFWGTLIT 123
V AK+ Y AFVYF+ A ++ Y I+ L +L K + W +I
Sbjct: 198 PVPWPAKFRYSPAFVYFVAALSVTGLYSIITTLASLFASNKPALKTKLLLYFILWDAVIN 377
Query: 124 IFDA-------------------------------------LMVALLFSGNGAATAIGVL 146
+ L++ ++ S G A + L
Sbjct: 378 PINRFFHLAIHLLFSN*LALIYIYQHESITNE*ISMMTLLQLILGIIASATGTAGGVAYL 557
Query: 147 GYQGNSHV-RWKKVCNVFDKYCHQVAASIILSQLGSLVFLLLVVL 190
G +GNSHV W K+C+V+DK+C V ASI ++ GS+V +LL+ L
Sbjct: 558 GLKGNSHVVGWNKICHVYDKFCRHVGASIAVALFGSIVTVLLIWL 692
>TC217892 similar to UP|Q6I7V6 (Q6I7V6) ATPase subunit 6, partial (10%)
Length = 989
Score = 73.2 bits (178), Expect = 8e-14
Identities = 39/155 (25%), Positives = 79/155 (50%)
Frame = +1
Query: 36 VLRFLGFVLTLAAAIVVGTDKQTTIVPIKVVDSLPPLNVAVSAKWHYLSAFVYFLVANAI 95
+LR + +L+ A+ V+ T+ QT ++ + A ++Y +F +F+ AN +
Sbjct: 232 ILRIIPTLLSAASIAVMVTNNQTVLI----------FAIRFQAHFYYSPSFKFFVAANGV 381
Query: 96 ACTYGAISMLLTLLNRGKSKVFWGTLITIFDALMVALLFSGNGAATAIGVLGYQGNSHVR 155
++++L L + ++ + + + D +M+ LL +G AATAIG +G G HV
Sbjct: 382 VVALSLLTLVLNFLMKRQASPRYHFFLFLHDIVMMVLLIAGCAAATAIGYVGQFGEDHVG 561
Query: 156 WKKVCNVFDKYCHQVAASIILSQLGSLVFLLLVVL 190
W+ +C+ K+C S++LS + + + +L
Sbjct: 562 WQPICDHVRKFCTTNLVSLLLSYFAFISYFGITLL 666
>TC233744
Length = 488
Score = 73.2 bits (178), Expect = 8e-14
Identities = 40/109 (36%), Positives = 66/109 (59%), Gaps = 1/109 (0%)
Frame = +3
Query: 88 YFLVANAIACTYGAISMLLTLLN-RGKSKVFWGTLITIFDALMVALLFSGNGAATAIGVL 146
+F++ NAIA Y + + + +L + K LI I D + +AL +G+GAAT + L
Sbjct: 6 FFVIVNAIASFYNMVVIGVEILGPQYDYKELRLGLIAILDVMTMALAATGDGAATFMAEL 185
Query: 147 GYQGNSHVRWKKVCNVFDKYCHQVAASIILSQLGSLVFLLLVVLLPILR 195
G GNSH RW K+C+ F+ YC++ ++I S +G L+ LL+V ++ I +
Sbjct: 186 GRNGNSHARWDKICDKFEAYCNRGGVALIASFVG-LILLLVVTVMSITK 329
>BI972961
Length = 396
Score = 66.2 bits (160), Expect = 9e-12
Identities = 37/104 (35%), Positives = 60/104 (57%), Gaps = 3/104 (2%)
Frame = +1
Query: 90 LVANAIACTYGAISMLLTLLNRGKSKVFWGTLI--TIFDALMVALLFSGNGAATAIGVLG 147
L A ++ Y I+ L +L K + L+ ++DAL++ ++ S G A + LG
Sbjct: 85 LSALSVTGLYSIITTLASLFASNKPALKTKLLLYFILWDALILGIIASATGTAGGVAYLG 264
Query: 148 YQGNSHV-RWKKVCNVFDKYCHQVAASIILSQLGSLVFLLLVVL 190
+GN HV W K+C+V+DK+C V ASI ++ GS+V +LL+ L
Sbjct: 265 LKGNRHVVGWNKICHVYDKFCRHVGASIAVALFGSVVTVLLIWL 396
>TC222694 weakly similar to GB|AAG40073.2|13265596|AF324722 AT4g15610
{Arabidopsis thaliana;} , partial (39%)
Length = 690
Score = 65.5 bits (158), Expect = 2e-11
Identities = 28/63 (44%), Positives = 44/63 (69%)
Frame = +2
Query: 128 LMVALLFSGNGAATAIGVLGYQGNSHVRWKKVCNVFDKYCHQVAASIILSQLGSLVFLLL 187
L++ + S GAA ++ +G +GNSHV W KVCN++DK+C +A SI ++ GS+V +LL
Sbjct: 359 LILGITASATGAAGSVAYIGLKGNSHVGWIKVCNIYDKFCRHLAGSIAVALFGSIVTVLL 538
Query: 188 VVL 190
+ L
Sbjct: 539 IWL 547
Score = 34.3 bits (77), Expect = 0.039
Identities = 33/116 (28%), Positives = 54/116 (46%), Gaps = 10/116 (8%)
Frame = +1
Query: 35 LVLRFLGFVLTLAAAIVVGTDKQTTIVPIKVVDSLPPLNVAVSAKWHYLSAFVYFLVANA 94
++LRFL ++ A V+ + QT V + V P AK+ Y AFVYF+ A +
Sbjct: 1 VILRFLLLAASVVALAVIVSSDQTEQVLFQDVLLPQP------AKFKYSPAFVYFVAAFS 162
Query: 95 IACTYGAISML--LTLLNRGKSK-------VFWGTLITIFDALMVAL-LFSGNGAA 140
++ Y +S L ++++ + K +FW +I L L +FS N A
Sbjct: 163 VSGLYALVSALASISVIQKPGFKLKFLLHFIFWDAVINQLAFLSTFLFIFSLNSLA 330
>TC209929 weakly similar to GB|AAM70533.1|21700819|AY124824
At2g35760/T20F21.5 {Arabidopsis thaliana;} , partial
(74%)
Length = 1070
Score = 61.2 bits (147), Expect = 3e-10
Identities = 47/158 (29%), Positives = 76/158 (47%), Gaps = 4/158 (2%)
Frame = +1
Query: 34 ELVLRFLGFVLTLAAAIVVGTDKQTTIVPIKVVDSLPPLNVAVSAKWHYLSAFVYFLVAN 93
ELVLR + L A ++V TD Q +K S AK+ + A V+ +VAN
Sbjct: 487 ELVLRCVSLGLGAVAIVLVVTDSQ-----VKEFFSFQK-----KAKFTDMKALVFLVVAN 636
Query: 94 AIACTYGAISMLLTLLNRGKSKVFWGT----LITIFDALMVALLFSGNGAATAIGVLGYQ 149
+ Y I L +++ + V + LI D +M + + AA GVLG
Sbjct: 637 GLTVGYSLIQGLRCVVSMIRGNVLFNKPLAWLIFSGDQVMAYVTVAAVAAALQSGVLGRT 816
Query: 150 GNSHVRWKKVCNVFDKYCHQVAASIILSQLGSLVFLLL 187
G + ++W KVCN++ K+C+Q+ I + + SL ++L
Sbjct: 817 GQAELQWMKVCNMYGKFCNQMGEGIASAFVVSLSKVVL 930
>TC219282 weakly similar to UP|Q9FFZ7 (Q9FFZ7) Gb|AAF00668.1, partial (82%)
Length = 751
Score = 59.3 bits (142), Expect = 1e-09
Identities = 45/193 (23%), Positives = 82/193 (42%), Gaps = 9/193 (4%)
Frame = +1
Query: 15 NYGRDELAMKKPPAGGG------SCELVLRFLGFVLTLAAAIVVGTDKQTTIVPIKVVDS 68
N + + + PP GG + +LR L AA +GT QT
Sbjct: 85 NVAKGKAVLAAPPRPGGWKKGVAIMDFILRLGAIAAALGAAATMGTSDQT---------- 234
Query: 69 LPPLN--VAVSAKWHYLSAFVYFLVANAIACTYGAISMLLTLLNRGKSKVFWGTL-ITIF 125
LP A + ++F +F++ A+ Y +S+ + + + L + I
Sbjct: 235 LPFFTQFFQFEASYDSFTSFQFFVITMALVGGYLVLSLPFSFVAIIRPHAAGPRLFLIIL 414
Query: 126 DALMVALLFSGNGAATAIGVLGYQGNSHVRWKKVCNVFDKYCHQVAASIILSQLGSLVFL 185
D + + L + +A AI L + GN W +CN F +C Q +++++ S + +V +
Sbjct: 415 DTVFLTLATASGASAAAIVYLAHNGNQDSNWLAICNQFGDFCAQTSSAVVSSFVAVVVLV 594
Query: 186 LLVVLLPILRSRR 198
LLVV+ + +R
Sbjct: 595 LLVVMSALSLGKR 633
>AW620830
Length = 430
Score = 55.8 bits (133), Expect = 1e-08
Identities = 40/139 (28%), Positives = 68/139 (48%), Gaps = 6/139 (4%)
Frame = +2
Query: 20 ELAMKKPPAGG-----GSCELVLRFLGFVLTLAAAIVVGTDKQTTIVPIKVVDSLPPLNV 74
E++ PP G + +LR + + TL +A+ +GT +QT + V
Sbjct: 38 EISKGAPPRKGLIRGLSIMDFILRIVAAIATLGSALGMGTTRQTLPFSTQFVK------- 196
Query: 75 AVSAKWHYLSAFVYFLVANAIACTYGAISMLLTLLNRGKSKVFWGTLITIF-DALMVALL 133
A + L FV+F+ +N+I C Y +S++L+ + +S ++ +F D +M LL
Sbjct: 197 -FRAVFSDLPTFVFFVTSNSIVCGYLVLSLVLSFFHIVRSAAVKSKVLQVFLDTVMYGLL 373
Query: 134 FSGNGAATAIGVLGYQGNS 152
+G AATAI + GNS
Sbjct: 374 TTGASAATAIVYEAHYGNS 430
>BI699872
Length = 420
Score = 52.8 bits (125), Expect = 1e-07
Identities = 31/114 (27%), Positives = 59/114 (51%), Gaps = 4/114 (3%)
Frame = +2
Query: 78 AKWHYLSAFVYFLVANAIACTYGAISMLLTLLNRGKSKVFWGTLIT--IF--DALMVALL 133
AK+ + + V+ +VAN +A Y I L +L+ + +V + + IF D +M +
Sbjct: 23 AKFTDMKSLVFLVVANGLAAGYSLIQGLRCILSMIRGRVLFSKPLAWAIFSGDQVMAYVT 202
Query: 134 FSGNGAATAIGVLGYQGNSHVRWKKVCNVFDKYCHQVAASIILSQLGSLVFLLL 187
AA G++ G ++W K+CN++ K+C+QV I + + SL +++
Sbjct: 203 VVALAAAGQSGMIARVGQPELQWMKICNMYGKFCNQVGEGIASAFVASLSMVVM 364
>BF069934
Length = 388
Score = 51.2 bits (121), Expect = 3e-07
Identities = 25/35 (71%), Positives = 28/35 (79%)
Frame = +2
Query: 88 YFLVANAIACTYGAISMLLTLLNRGKSKVFWGTLI 122
YFLV NAIACTY +S+LL L+NRGKSK W TLI
Sbjct: 284 YFLVTNAIACTYAVLSLLLALVNRGKSKGLW-TLI 385
>TC218814
Length = 881
Score = 49.7 bits (117), Expect = 9e-07
Identities = 38/160 (23%), Positives = 69/160 (42%), Gaps = 6/160 (3%)
Frame = +3
Query: 34 ELVLRFLGFVLTLAAAIVVGTDKQTTIVPIKVVDSLPPLNVAVSAKWHYLSAFVYFLVAN 93
E+ LR ++ + A +V D QT +V + + A + L VY A+
Sbjct: 153 EVYLRVSAILVLVLTACLVALDTQTKVVFLSIEKK------ATCRDLNALKILVYVTSAS 314
Query: 94 AIACTYGAISMLLTLLNRGKSKVFWGTLIT------IFDALMVALLFSGNGAATAIGVLG 147
A G +LL + K F G+ + + D + V + F+ N AA +L
Sbjct: 315 A-----GYNLLLLCKHSIWSRKNFKGSYLCMAWICFLLDQIAVYMTFAANTAAMGAAMLA 479
Query: 148 YQGNSHVRWKKVCNVFDKYCHQVAASIILSQLGSLVFLLL 187
G+ +W KVC+ F ++C Q+ +++ S++ L+
Sbjct: 480 ITGSDAFQWLKVCDKFTRFCVQIGGALLCGYAASIIMALI 599
>TC216187
Length = 977
Score = 48.1 bits (113), Expect = 3e-06
Identities = 25/114 (21%), Positives = 53/114 (45%)
Frame = +1
Query: 77 SAKWHYLSAFVYFLVANAIACTYGAISMLLTLLNRGKSKVFWGTLITIFDALMVALLFSG 136
S + L AF Y + AN I Y S ++ + S + + D ++ ++ +
Sbjct: 304 SVDYSDLGAFRYLVHANGICAGYSLFSAVIAAMPC-PSTIPRAWTFFLLDQVLTYIILAA 480
Query: 137 NGAATAIGVLGYQGNSHVRWKKVCNVFDKYCHQVAASIILSQLGSLVFLLLVVL 190
+T + L +G++ W C F ++CH+V AS+ ++ + ++LL ++
Sbjct: 481 GAVSTEVLYLAEKGDAATTWSSACGSFGRFCHKVTASVAITFVAVFCYVLLSLI 642
>TC216185
Length = 1098
Score = 47.4 bits (111), Expect = 4e-06
Identities = 25/111 (22%), Positives = 50/111 (44%)
Frame = +1
Query: 77 SAKWHYLSAFVYFLVANAIACTYGAISMLLTLLNRGKSKVFWGTLITIFDALMVALLFSG 136
S + L AF Y + AN I Y S ++ + S + + D ++ ++ +
Sbjct: 403 SVDYSDLGAFRYLVHANGICAGYSLFSAVIAAMPC-PSTIPRAWTFFLLDQVLTYIILAA 579
Query: 137 NGAATAIGVLGYQGNSHVRWKKVCNVFDKYCHQVAASIILSQLGSLVFLLL 187
+T + L G++ W C F ++CH+V AS+ ++ + ++LL
Sbjct: 580 GAVSTEVLYLAENGDAATTWSSACGSFGRFCHKVTASVAITFVAVFCYVLL 732
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.326 0.141 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,627,151
Number of Sequences: 63676
Number of extensions: 130023
Number of successful extensions: 954
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 941
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 943
length of query: 200
length of database: 12,639,632
effective HSP length: 92
effective length of query: 108
effective length of database: 6,781,440
effective search space: 732395520
effective search space used: 732395520
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 56 (26.2 bits)
Medicago: description of AC147364.1


