

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147363.5 - phase: 0
(433 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
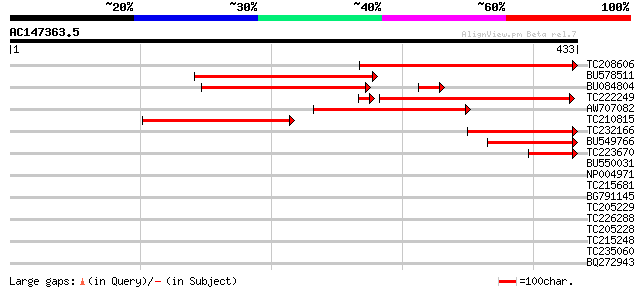
TC208606 similar to PIR|T02655|T02655 hydroxymethylglutaryl-CoA ... 315 3e-86
BU578511 265 3e-71
BU084804 225 3e-64
TC222249 similar to PIR|T02655|T02655 hydroxymethylglutaryl-CoA ... 221 2e-60
AW707082 145 3e-35
TC210815 similar to PIR|T02655|T02655 hydroxymethylglutaryl-CoA ... 144 1e-34
TC232166 similar to PIR|T02655|T02655 hydroxymethylglutaryl-CoA ... 140 1e-33
BU549766 107 1e-23
TC223670 58 7e-09
BU550031 39 0.005
NP004971 soybean late nodulin 37 0.013
TC215681 similar to UP|Q9LPR4 (Q9LPR4) F15H18.3, partial (79%) 32 0.43
BG791145 32 0.73
TC205229 similar to UP|Q8W0Z9 (Q8W0Z9) AT3g58560/F14P22_150, par... 32 0.73
TC226288 similar to UP|RK13_SPIOL (P12629) 50S ribosomal protein... 30 2.8
TC205228 29 3.6
TC215248 weakly similar to UP|Q94II4 (Q94II4) NAM-like protein, ... 28 6.2
TC235060 weakly similar to UP|O64785 (O64785) T1F9.16 (At1g61350... 28 8.0
BQ272943 28 8.0
>TC208606 similar to PIR|T02655|T02655 hydroxymethylglutaryl-CoA lyase -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(36%)
Length = 1157
Score = 315 bits (807), Expect = 3e-86
Identities = 154/166 (92%), Positives = 161/166 (96%)
Frame = +1
Query: 268 YVSCVVGCPVEGPVPPSKVAYVAKALYDMGCFEISLGDTIGVGTPGTVVPMLLAVMAIVP 327
YVSCVVGCPVEGP+PPSKVAYVAK LYDMGCFEISLGDTIGVGTPGTVVPMLLAVMA+VP
Sbjct: 295 YVSCVVGCPVEGPIPPSKVAYVAKELYDMGCFEISLGDTIGVGTPGTVVPMLLAVMAVVP 474
Query: 328 TEKLAVHFHDTYGQSLPNILVSLQMGISAVDSSVAGLGGCPYAKGASGNVATEDVVYMLN 387
EKLAVHFHDTYGQSLPNILVSLQMGISAVDSSVAGLGGCPYAKGASGNVATEDVVYMLN
Sbjct: 475 AEKLAVHFHDTYGQSLPNILVSLQMGISAVDSSVAGLGGCPYAKGASGNVATEDVVYMLN 654
Query: 388 GLGIKTNVDIGKLMSAGDFIGKQLGRPSGSKTAIALNRVTADASKI 433
GLG+KTNVD+GKLM AG+FIGK LGRPSGSKTA+A +RVTADASKI
Sbjct: 655 GLGVKTNVDLGKLMLAGEFIGKHLGRPSGSKTAVAFSRVTADASKI 792
>BU578511
Length = 423
Score = 265 bits (677), Expect = 3e-71
Identities = 134/140 (95%), Positives = 138/140 (97%)
Frame = +3
Query: 142 RDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQLADAKDVMQAVHNLRG 201
RDGLQNEKNIVPTDVKIELIHRLAS+GLSVIEATSFVSPKWVPQLADAKDVMQAVHNL G
Sbjct: 3 RDGLQNEKNIVPTDVKIELIHRLASSGLSVIEATSFVSPKWVPQLADAKDVMQAVHNLGG 182
Query: 202 IRLPVLTPNLKGFEAAVAAGAREVAVFASASESFSKSNINCSIEESLSRYRAVTRAAKEL 261
IRLPVLTPNLKGFEAA+AAGAREVAVFASASESFSKSNINCSIEESL RYRAVTRAAK+L
Sbjct: 183 IRLPVLTPNLKGFEAAIAAGAREVAVFASASESFSKSNINCSIEESLVRYRAVTRAAKQL 362
Query: 262 SIPVRGYVSCVVGCPVEGPV 281
SIPVRGYVSCVVGCPVEGP+
Sbjct: 363 SIPVRGYVSCVVGCPVEGPI 422
>BU084804
Length = 424
Score = 225 bits (574), Expect(2) = 3e-64
Identities = 119/130 (91%), Positives = 122/130 (93%), Gaps = 1/130 (0%)
Frame = +3
Query: 147 NEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQLADAKDVMQAVHNLRGIRLPV 206
NEKNIVPTDVKIELIHRLAS+GLSVIEATSFVSPKWVPQLADAKDVMQAVHNL GIRLPV
Sbjct: 3 NEKNIVPTDVKIELIHRLASSGLSVIEATSFVSPKWVPQLADAKDVMQAVHNLGGIRLPV 182
Query: 207 LTPNLKGFEAAVAAGAREVAVFASASESFSKSNINCSIEESLSRYRAVTRAAKELSIPVR 266
LTPNLKGFEAA+AAGAREVAVFASASESFSKSNINCSIEESL RYRAVTRAAK+LSIPVR
Sbjct: 183 LTPNLKGFEAAIAAGAREVAVFASASESFSKSNINCSIEESLVRYRAVTRAAKQLSIPVR 362
Query: 267 GYVSC-VVGC 275
G S GC
Sbjct: 363 GNCSSNAFGC 392
Score = 38.5 bits (88), Expect(2) = 3e-64
Identities = 18/20 (90%), Positives = 19/20 (95%)
Frame = +1
Query: 313 GTVVPMLLAVMAIVPTEKLA 332
GTVVPMLLAVMA+VP EKLA
Sbjct: 364 GTVVPMLLAVMAVVPAEKLA 423
>TC222249 similar to PIR|T02655|T02655 hydroxymethylglutaryl-CoA lyase -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(34%)
Length = 493
Score = 221 bits (564), Expect(2) = 2e-60
Identities = 108/149 (72%), Positives = 126/149 (84%)
Frame = +2
Query: 283 PSKVAYVAKALYDMGCFEISLGDTIGVGTPGTVVPMLLAVMAIVPTEKLAVHFHDTYGQS 342
P+K+AYVAK+LYDMGC EISLGDTIGVGTPGT + ML AV+ +VPT+ LA HFHDTYGQ+
Sbjct: 47 PAKIAYVAKSLYDMGCSEISLGDTIGVGTPGTDISMLEAVLDVVPTDMLADHFHDTYGQA 226
Query: 343 LPNILVSLQMGISAVDSSVAGLGGCPYAKGASGNVATEDVVYMLNGLGIKTNVDIGKLMS 402
L NIL+SLQMGIS VDSS +GL GCPYAKGA+GNV T+DVVYMLNGLG+ TNVD+ KLM
Sbjct: 227 LSNILISLQMGISTVDSSASGLEGCPYAKGATGNVTTDDVVYMLNGLGVNTNVDLAKLMR 406
Query: 403 AGDFIGKQLGRPSGSKTAIALNRVTADAS 431
AGDFI K LGR SGS+ ++RVT+ AS
Sbjct: 407 AGDFICKHLGRASGSEAGTHMSRVTSHAS 493
Score = 29.6 bits (65), Expect(2) = 2e-60
Identities = 10/12 (83%), Positives = 12/12 (99%)
Frame = +1
Query: 267 GYVSCVVGCPVE 278
GY+SCVVGCP+E
Sbjct: 1 GYISCVVGCPLE 36
>AW707082
Length = 362
Score = 145 bits (366), Expect = 3e-35
Identities = 67/120 (55%), Positives = 94/120 (77%)
Frame = +3
Query: 233 ESFSKSNINCSIEESLSRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKA 292
+SFSKSNIN SIEE L Y+A+T+ K LSIP++ Y+S ++ PV+ P+PPSK+ Y++K
Sbjct: 3 KSFSKSNINYSIEEILVHYQAITQTTKHLSIPIQKYISYIIKYPVKRPIPPSKLTYISKE 182
Query: 293 LYDMGCFEISLGDTIGVGTPGTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQM 352
LY++ F+ISL +TI V TP T+VP+LL ++ I+P E L +HFH+TY QSLPNIL+SLQ+
Sbjct: 183 LYNINYFKISLKNTIKVNTPKTIVPILLTIITIMPAENLTIHFHNTYKQSLPNILISLQI 362
>TC210815 similar to PIR|T02655|T02655 hydroxymethylglutaryl-CoA lyase -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(20%)
Length = 634
Score = 144 bits (362), Expect = 1e-34
Identities = 70/116 (60%), Positives = 89/116 (76%)
Frame = +2
Query: 102 SFYTSDYQYSQKRNNKDMQDMAYKFMKGMPEFVKIVEVGPRDGLQNEKNIVPTDVKIELI 161
S Y + ++ N+ ++ + K + +P++VKIVEVG RDGLQNEK I+PT+VK+ELI
Sbjct: 287 SCYVVNRHFASDCNDICSKEFSSKLLTSIPDYVKIVEVGARDGLQNEKAIIPTNVKVELI 466
Query: 162 HRLASTGLSVIEATSFVSPKWVPQLADAKDVMQAVHNLRGIRLPVLTPNLKGFEAA 217
L S+GLSV+E TSFVSPKWVPQLADAKDV+ A+ N+ G R PVLTPNLKGFEAA
Sbjct: 467 KLLVSSGLSVVETTSFVSPKWVPQLADAKDVLAAIQNVEGARFPVLTPNLKGFEAA 634
>TC232166 similar to PIR|T02655|T02655 hydroxymethylglutaryl-CoA lyase -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(18%)
Length = 447
Score = 140 bits (353), Expect = 1e-33
Identities = 71/84 (84%), Positives = 77/84 (91%)
Frame = +2
Query: 350 LQMGISAVDSSVAGLGGCPYAKGASGNVATEDVVYMLNGLGIKTNVDIGKLMSAGDFIGK 409
L MGIS VDSSVAGLGGCPYAKGASGNVATEDVVYMLNGLGIKT+VD+GKL+ AG+FI
Sbjct: 74 LLMGISTVDSSVAGLGGCPYAKGASGNVATEDVVYMLNGLGIKTDVDLGKLILAGEFISN 253
Query: 410 QLGRPSGSKTAIALNRVTADASKI 433
LGRPS SKTAIALNRVT++ASKI
Sbjct: 254 HLGRPSTSKTAIALNRVTSNASKI 325
>BU549766
Length = 458
Score = 107 bits (267), Expect = 1e-23
Identities = 52/68 (76%), Positives = 58/68 (84%)
Frame = -1
Query: 366 GCPYAKGASGNVATEDVVYMLNGLGIKTNVDIGKLMSAGDFIGKQLGRPSGSKTAIALNR 425
GCPY KGA NVATEDVVYMLNGLG+KTNVD+GKLM AGDFI K LGR SGSK A AL++
Sbjct: 458 GCPYXKGAXXNVATEDVVYMLNGLGVKTNVDLGKLMLAGDFICKHLGRASGSKAATALSK 279
Query: 426 VTADASKI 433
VT+ ASK+
Sbjct: 278 VTSHASKL 255
>TC223670
Length = 409
Score = 58.2 bits (139), Expect = 7e-09
Identities = 29/37 (78%), Positives = 32/37 (86%)
Frame = +3
Query: 397 IGKLMSAGDFIGKQLGRPSGSKTAIALNRVTADASKI 433
+GKLM AGDFI LGRPS SKTAIALNRVT++ASKI
Sbjct: 3 LGKLMLAGDFISNHLGRPSTSKTAIALNRVTSNASKI 113
>BU550031
Length = 579
Score = 38.9 bits (89), Expect = 0.005
Identities = 23/97 (23%), Positives = 47/97 (47%), Gaps = 3/97 (3%)
Frame = -1
Query: 297 GCFEISLGDTIGVGTPGTVVPMLLAVMAIVPTEK---LAVHFHDTYGQSLPNILVSLQMG 353
G + +GDT+G+ P + ++ + A VP + +++H H+ + N + + Q G
Sbjct: 495 GATTLGIGDTVGITMPFEIRQLIADIKANVPGAENVIISMHCHNDXXHATANAIEAAQAG 316
Query: 354 ISAVDSSVAGLGGCPYAKGASGNVATEDVVYMLNGLG 390
++ ++ G+G +GN + E+VV L G
Sbjct: 315 AMQLEVTINGIG------ERAGNASLEEVVMALKCRG 223
>NP004971 soybean late nodulin
Length = 1698
Score = 37.4 bits (85), Expect = 0.013
Identities = 51/281 (18%), Positives = 112/281 (39%), Gaps = 19/281 (6%)
Frame = +1
Query: 133 FVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEA-------TSFVSPKWVPQ 185
+V+I++ RDG Q+ + K+++ +L G+ +I+ + F++ K + Q
Sbjct: 94 YVRILDTTLRDGEQSPGATMTAKEKLDIARQLVKLGVDIIQPGFPSASNSDFMAVKMIAQ 273
Query: 186 -LADAKDVMQAVHNLRGIRLPVLTPNLKGFEAAVAAGAREVAVFASASESFSKSNINCSI 244
+ +A D V + G V +EA A + + S + + S
Sbjct: 274 EVGNAVDDDGYVPVIAGFCRCVEKDISTAWEAVKYAKRPRLCTSIATSPIHMEHKLRKSK 453
Query: 245 EESLSRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKALYDMGCFEISLG 304
++ + R + + A+ L G G + + + + G +++
Sbjct: 454 DQVIQIARDMVKFARSL-----GCNDIQFGAEDATRSDREFLYEILGVVIEAGATTVNIA 618
Query: 305 DTIGVGTPGTVVPMLLAVMAIVP---TEKLAVHFHDTYGQSLPNILVSLQMGISAVDSSV 361
DT+G+ P + +++ + P ++ H H+ G + N + + G ++ ++
Sbjct: 619 DTVGIVMPLELGKLIVDIKDNTPGIANVIISTHCHNDLGLATANTIEGARTGARQLEVTI 798
Query: 362 AGLGGCPYAKGASGNVATEDVV--------YMLNGLGIKTN 394
G+G +GN + E+VV + LNGL + N
Sbjct: 799 NGIG------ERAGNASLEEVVMALASKGDHALNGLYTRIN 903
>TC215681 similar to UP|Q9LPR4 (Q9LPR4) F15H18.3, partial (79%)
Length = 2427
Score = 32.3 bits (72), Expect = 0.43
Identities = 45/235 (19%), Positives = 93/235 (39%), Gaps = 12/235 (5%)
Frame = +3
Query: 131 PEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEA-------TSFVSPKWV 183
P +V+I + RDG Q+ + + K+++ +LA G+ +IEA F + K +
Sbjct: 270 PSYVRIFDTTLRDGEQSPGASMTSKEKLDVARQLAKLGVDIIEAGFPAASKDDFEAVKMI 449
Query: 184 PQ-LADAKDVMQAVHNLRGIRLPVLTPNLKGFEAAVAAGAREVAVFASASESFSKSNINC 242
Q + +A D V + G+ +EA A + F + S + +
Sbjct: 450 AQEVGNAVDDDGYVPVICGLSRCNEKDIRTAWEAVKYAKRPRIHTFIATSAIHMEYKLRM 629
Query: 243 SIEESLSRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAY-VAKALYDMGCFEI 301
S ++ + R + + A+ L V P + + Y + + +G +
Sbjct: 630 SKDKVVDIARNMVKFARSLG------CDDVEFSPEDAGRSDREFLYEILGEVIKVGATTL 791
Query: 302 SLGDTIGVGTPGTVVPMLLAVMAIVPTEK---LAVHFHDTYGQSLPNILVSLQMG 353
++ DT+G+ P ++ + A P + ++ H + G S N + + G
Sbjct: 792 NIPDTVGITMPSEFGKLIADIKANTPGIENVIISTHCQNDLGLSTANTIEGARAG 956
>BG791145
Length = 401
Score = 31.6 bits (70), Expect = 0.73
Identities = 11/35 (31%), Positives = 18/35 (51%)
Frame = +1
Query: 22 VQRFSSGCCRPQVDNLGMGNCFIEGRSCSTSNSCN 56
++R CC+ + N+G+ R CS+ SCN
Sbjct: 262 LERIGENCCKCRYSNVGLWGL*FNSRKCSSCTSCN 366
>TC205229 similar to UP|Q8W0Z9 (Q8W0Z9) AT3g58560/F14P22_150, partial (57%)
Length = 1743
Score = 31.6 bits (70), Expect = 0.73
Identities = 11/35 (31%), Positives = 18/35 (51%)
Frame = +3
Query: 22 VQRFSSGCCRPQVDNLGMGNCFIEGRSCSTSNSCN 56
++R CC+ + N+G+ R CS+ SCN
Sbjct: 594 LERIGENCCKCRYSNVGLWGL*FNSRKCSSCTSCN 698
>TC226288 similar to UP|RK13_SPIOL (P12629) 50S ribosomal protein L13,
chloroplast precursor (CL13), partial (62%)
Length = 1054
Score = 29.6 bits (65), Expect = 2.8
Identities = 12/33 (36%), Positives = 18/33 (54%)
Frame = -2
Query: 196 VHNLRGIRLPVLTPNLKGFEAAVAAGAREVAVF 228
+H + +P L PN++ FE+ ARE VF
Sbjct: 360 IHMICSFWIPALVPNVRAFESGFGFDAREARVF 262
>TC205228
Length = 795
Score = 29.3 bits (64), Expect = 3.6
Identities = 11/33 (33%), Positives = 16/33 (48%)
Frame = +3
Query: 24 RFSSGCCRPQVDNLGMGNCFIEGRSCSTSNSCN 56
R CC+ + N+G+ R CS+ SCN
Sbjct: 48 RIGENCCQCRHSNVGLWGL*FNSRKCSSCTSCN 146
>TC215248 weakly similar to UP|Q94II4 (Q94II4) NAM-like protein, partial
(15%)
Length = 911
Score = 28.5 bits (62), Expect = 6.2
Identities = 16/39 (41%), Positives = 21/39 (53%), Gaps = 2/39 (5%)
Frame = -2
Query: 25 FSSGCCRPQVDNLGMGN--CFIEGRSCSTSNSCNEDNED 61
FSS CC V G+G I G TS+SC+E+ E+
Sbjct: 193 FSSCCCCCSVLTRGIGKHRSSIAGSDSRTSSSCDEEEEE 77
>TC235060 weakly similar to UP|O64785 (O64785) T1F9.16 (At1g61350/T1F9_16),
partial (16%)
Length = 455
Score = 28.1 bits (61), Expect = 8.0
Identities = 12/33 (36%), Positives = 17/33 (51%)
Frame = +3
Query: 23 QRFSSGCCRPQVDNLGMGNCFIEGRSCSTSNSC 55
Q+ G CR L NC+I+GR+ + SC
Sbjct: 84 QKIGRGLCRMTKTLLFFCNCWIQGRAIQVTKSC 182
>BQ272943
Length = 420
Score = 28.1 bits (61), Expect = 8.0
Identities = 11/21 (52%), Positives = 13/21 (61%)
Frame = +3
Query: 68 PWKRQTRDMSRGDSFSPRTMT 88
PW +TR M+ S PRTMT
Sbjct: 150 PWCTRTRSMTSSSSLEPRTMT 212
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.133 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,915,106
Number of Sequences: 63676
Number of extensions: 221114
Number of successful extensions: 1038
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 1032
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1037
length of query: 433
length of database: 12,639,632
effective HSP length: 100
effective length of query: 333
effective length of database: 6,272,032
effective search space: 2088586656
effective search space used: 2088586656
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC147363.5


