

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147202.9 + phase: 0 /pseudo
(520 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
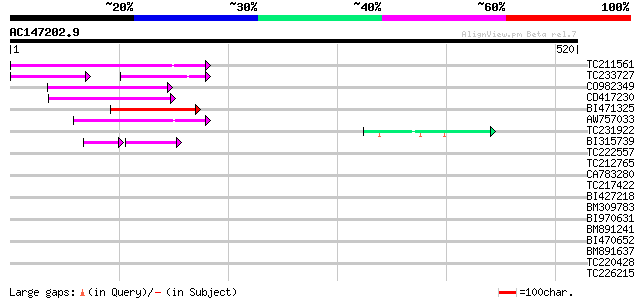
Score E
Sequences producing significant alignments: (bits) Value
TC211561 weakly similar to UP|O22675 (O22675) Reverse transcript... 109 3e-24
TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR ret... 54 2e-16
CO982349 77 2e-14
CD417230 weakly similar to GP|2244915|emb| reverse transcriptase... 74 2e-13
BI471325 67 2e-11
AW757033 67 3e-11
TC231922 45 1e-04
BI315739 35 4e-04
TC222557 40 0.003
TC212765 39 0.007
CA783280 weakly similar to PIR|T01871|T01 RNA-directed DNA polym... 38 0.012
TC217422 UP|O24422 (O24422) Desiccation protective protein LEA5,... 36 0.047
BI427218 34 0.14
BM309783 33 0.40
BI970631 32 0.68
BM891241 32 0.89
BI470652 similar to PIR|T17102|T171 RNA-directed DNA polymerase ... 29 4.4
BM891637 29 4.4
TC220428 weakly similar to UP|Q9FI28 (Q9FI28) Laccase (Diphenol ... 29 5.8
TC226215 homologue to UP|Q6S4R9 (Q6S4R9) Allantoinase , partial... 28 7.6
>TC211561 weakly similar to UP|O22675 (O22675) Reverse transcriptase
(Fragment) , partial (59%)
Length = 1077
Score = 109 bits (273), Expect = 3e-24
Identities = 68/184 (36%), Positives = 90/184 (47%)
Frame = +1
Query: 1 LILKVDMSKAYDRINWEYLRRVMLRLGFDAKWFGWIMMCVESVKYKVLVNQESEGPILPG 60
L+ KVD KAYD ++W +L +M R+ F KW WI CV+S VLVN LP
Sbjct: 64 LVFKVDYEKAYDSVSWNFLMYMMWRMDFSPKWIKWIEECVKSASISVLVNGSPTAEFLPQ 243
Query: 61 SGLRKGDPLSPYLFILCAQGLTSLIDKAEGRGDLHGVRVCTGAPTFTHLLFVDDCFLFCR 120
GLR+GDPL+P+LF + A+GL L+ +A V + L + DD F
Sbjct: 244 RGLRQGDPLTPFLFNIVAEGLNGLMRRAVEENLYKPYLVGANGVPISILQYADDTIFFGE 423
Query: 121 AT*REVNALKNILSIYEDASGQQINFQKSEIFFSKNVGA*AKVNILNILQVNIAI*TGKY 180
A V A+K IL +E S +INF KS F V K N L ++ Y
Sbjct: 424 AAKENVEAIKVILRSFELVSNLRINFAKS-CFGVFGVTDQWKQEAANYLHCSLLAFPFIY 600
Query: 181 LGLP 184
LG+P
Sbjct: 601 LGIP 612
>TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR retroelement
reverse transcriptase, partial (7%)
Length = 956
Score = 54.3 bits (129), Expect(2) = 2e-16
Identities = 27/74 (36%), Positives = 44/74 (58%)
Frame = -3
Query: 1 LILKVDMSKAYDRINWEYLRRVMLRLGFDAKWFGWIMMCVESVKYKVLVNQESEGPILPG 60
L LK+D+ KA+D ++ ++L V+ + G+ + WI + + S K + VN E G
Sbjct: 765 LALKIDIVKAFDTLDRDFLISVLNKFGYSHTFCNWIRVILLSAKLSISVNGEPVGFFSCQ 586
Query: 61 SGLRKGDPLSPYLF 74
G+R+GDPLSP L+
Sbjct: 585 RGVRQGDPLSPSLY 544
Score = 49.3 bits (116), Expect(2) = 2e-16
Identities = 29/83 (34%), Positives = 46/83 (54%)
Frame = -2
Query: 102 GAPTFTHLLFVDDCFLFCRAT*REVNALKNILSIYEDASGQQINFQKSEIFFSKNVGA*A 161
G+ T +H++ DD +FC+ T R V+ L N +Y+D+ GQ I+ KS+IF G
Sbjct: 466 GSLTRSHVMHADDIMIFCKGTSRNVHNLINFFQLYKDSFGQVISKHKSKIFIGSIPGQRL 287
Query: 162 KVNILNILQVNIAI*TGKYLGLP 184
+V I ++LQ + Y G+P
Sbjct: 286 RV-ISDLLQYFHSDIPFNYFGVP 221
>CO982349
Length = 795
Score = 76.6 bits (187), Expect = 2e-14
Identities = 43/115 (37%), Positives = 60/115 (51%)
Frame = +3
Query: 35 WIMMCVESVKYKVLVNQESEGPILPGSGLRKGDPLSPYLFILCAQGLTSLIDKAEGRGDL 94
WI C+ S +LVN+ P GLR+GDPL+P LF + A+GLT L+ +A R
Sbjct: 69 WIRGCLHSASVSILVNESPINEFSPQRGLRQGDPLAPLLFNIVAEGLTGLMREAVDRKRF 248
Query: 95 HGVRVCTGAPTFTHLLFVDDCFLFCRAT*REVNALKNILSIYEDASGQQINFQKS 149
+ V + L + DD F AT + +K IL +E ASG +INF +S
Sbjct: 249 NSFLVGKNKEPVSILQYADDTIFFEEATMENMKVIKTILRCFELASGLKINFAES 413
>CD417230 weakly similar to GP|2244915|emb| reverse transcriptase like
protein {Arabidopsis thaliana}, partial (7%)
Length = 688
Score = 73.6 bits (179), Expect = 2e-13
Identities = 37/117 (31%), Positives = 63/117 (53%)
Frame = -3
Query: 36 IMMCVESVKYKVLVNQESEGPILPGSGLRKGDPLSPYLFILCAQGLTSLIDKAEGRGDLH 95
+ C+ + +L N E P G+R+ DP++PYLF+LC + L+ LI+ +
Sbjct: 677 VWYCMSTTTMSLLWNGEVLDHFSPNIGVRQRDPIAPYLFVLCLERLSHLINLVMNKKL*R 498
Query: 96 GVRVCTGAPTFTHLLFVDDCFLFCRAT*REVNALKNILSIYEDASGQQINFQKSEIF 152
++V G +HL FVDD LF A +V+ + L ++ D+SG ++N K++ F
Sbjct: 497 SIQVNRGGLLISHLAFVDDLILFVEANMDQVDVINLTLELFSDSSGAKVNVDKTKSF 327
>BI471325
Length = 421
Score = 67.0 bits (162), Expect = 2e-11
Identities = 37/83 (44%), Positives = 52/83 (62%)
Frame = +2
Query: 93 DLHGVRVCTGAPTFTHLLFVDDCFLFCRAT*REVNALKNILSIYEDASGQQINFQKSEIF 152
D+H V+V G +HLL VD FLFCR +E + LKNIL+ +E +SG +N +KSEI
Sbjct: 5 DIHRVKVHRGITILSHLLSVDFYFLFCRTIDKEADVLKNILTTFEASSGLVVNLRKSEIC 184
Query: 153 FSKNVGA*AKVNILNILQVNIAI 175
FS+N A+ +I L ++I I
Sbjct: 185 FSRNTSQIARDSIFFHLGMSIWI 253
>AW757033
Length = 441
Score = 66.6 bits (161), Expect = 3e-11
Identities = 45/126 (35%), Positives = 63/126 (49%)
Frame = -2
Query: 59 PGSGLRKGDPLSPYLFILCAQGLTSLIDKAEGRGDLHGVRVCTGAPTFTHLLFVDDCFLF 118
P GLR+GDPL+P LF + A+GLT L+ +A R V + L + DD F
Sbjct: 425 PHRGLRQGDPLAPLLFNIAAEGLTGLMREAVARNHYKSFAVGKYKKPVSILQYADDTIFF 246
Query: 119 CRAT*REVNALKNILSIYEDASGQQINFQKSEIFFSKNVGA*AKVNILNILQVNIAI*TG 178
AT V +K IL +E ASG +INF KS F + +V * + + ++
Sbjct: 245 GEATMENVRVIKTILRGFELASGLKINFAKSR-FGAISVPD*WRKEAAEFMNCSLLSLPF 69
Query: 179 KYLGLP 184
YLG+P
Sbjct: 68 SYLGIP 51
>TC231922
Length = 803
Score = 44.7 bits (104), Expect = 1e-04
Identities = 31/139 (22%), Positives = 55/139 (39%), Gaps = 18/139 (12%)
Frame = -3
Query: 325 SYVWKSIHEAIPM-----VKQGTRWKIGNGDKIRVFQDRWLVNGGLAQPAA*YNRL---- 375
S+ WK + + Q W++G GDKI+ +QD WL G Q YN+L
Sbjct: 777 SHWWKDLRRLYNQPDFHSIHQNMVWRVGCGDKIKFWQDSWLSEGCNLQQK--YNQLYTIS 604
Query: 376 ----LQLNVKELLQPGQKSWNLDVLQ-----HYLSSQSVFEAMNTLLFNSTAYDERVWRP 426
L ++ +SW + Y + ++ + + A+D W+
Sbjct: 603 RQ*NLTISKMGKFSQNARSWEFKWRRRLFDYEYAMAVDFMNEISGISIQNQAHDSMFWKA 424
Query: 427 GKRGIYTVRSASRFCLTEL 445
G+Y+ +SA R + +
Sbjct: 423 DSSGVYSTKSAYRLLMPSI 367
>BI315739
Length = 442
Score = 35.0 bits (79), Expect(2) = 4e-04
Identities = 18/51 (35%), Positives = 28/51 (54%)
Frame = +2
Query: 107 THLLFVDDCFLFCRAT*REVNALKNILSIYEDASGQQINFQKSEIFFSKNV 157
+HL F D+C LF +AT +V ++++L + G + QKS KNV
Sbjct: 167 SHLFFPDNCLLFVKATSSQVKLVRDVLQQFCLT*GLKAGLQKSRFLA*KNV 319
Score = 26.9 bits (58), Expect(2) = 4e-04
Identities = 12/37 (32%), Positives = 17/37 (45%)
Frame = +3
Query: 68 PLSPYLFILCAQGLTSLIDKAEGRGDLHGVRVCTGAP 104
PLSPYLF +C + L LI +++ P
Sbjct: 48 PLSPYLF*ICMEKLAILIQSKVNEETWEPIKISRNGP 158
>TC222557
Length = 1002
Score = 40.0 bits (92), Expect = 0.003
Identities = 33/113 (29%), Positives = 54/113 (47%), Gaps = 14/113 (12%)
Frame = +3
Query: 344 WKIGNGDKIRVFQDRWLVNGGLAQP-AA*YNRLLQLNVKE---LLQPGQKS-----WNLD 394
WK+G GDKIR ++D WL+ +P A Y RL ++ ++ +L G S W L+
Sbjct: 69 WKVGCGDKIRFWEDCWLME---QEPLRAKYPRLYNISCQQQKLILVMGFHSANVWEWKLE 239
Query: 395 VLQHYLSSQ-----SVFEAMNTLLFNSTAYDERVWRPGKRGIYTVRSASRFCL 442
+H ++ S + ++ + D VW+P G ++ RSA R L
Sbjct: 240 WRRHLFDNEVQAAASFLDDISWGHIDRWTLDCWVWKPEPNGQFSTRSAYRMLL 398
>TC212765
Length = 637
Score = 38.5 bits (88), Expect = 0.007
Identities = 33/118 (27%), Positives = 48/118 (39%)
Frame = +3
Query: 71 PYLFILCAQGLTSLIDKAEGRGDLHGVRVCTGAPTFTHLLFVDDCFLFCRAT*REVNALK 130
PYLF+LC + L I + + V +L+ VDD FLF A
Sbjct: 15 PYLFVLCMERLALCIHDLTSQ*IWRPILVSDDNRLVPYLMLVDDIFLFINAIGD*AYLAS 194
Query: 131 NILSIYEDASGQQINFQKSEIFFSKNVGA*AKVNILNILQVNIAI*TGKYLGLPSVIG 188
L + G ++N KS ++ S + + I N L + A GKYL L + G
Sbjct: 195 LTL*NFFQTFGLKLNLDKSSMYCS*RMAPVLRDLISNTLNIVHAPSLGKYLCLKFIHG 368
>CA783280 weakly similar to PIR|T01871|T01 RNA-directed DNA polymerase
homolog T24M8.8 - Arabidopsis thaliana, partial (4%)
Length = 456
Score = 37.7 bits (86), Expect = 0.012
Identities = 20/61 (32%), Positives = 35/61 (56%), Gaps = 6/61 (9%)
Frame = +2
Query: 305 LKARYYRRNAIFEANKGNNLSYVWKSIHEA------IPMVKQGTRWKIGNGDKIRVFQDR 358
L+++Y A+ E + G+ S WK + + IP+ K+ T WK+G GD+I+ ++D
Sbjct: 242 LQSKYGGWRALEEGSSGSKDSAWWKDLIKTQQLQRNIPL-KRETIWKVGGGDRIKFWEDL 418
Query: 359 W 359
W
Sbjct: 419 W 421
>TC217422 UP|O24422 (O24422) Desiccation protective protein LEA5, complete
Length = 627
Score = 35.8 bits (81), Expect = 0.047
Identities = 14/30 (46%), Positives = 21/30 (69%)
Frame = +2
Query: 21 RVMLRLGFDAKWFGWIMMCVESVKYKVLVN 50
+++LR+ F WFG + +C S KYKVL+N
Sbjct: 476 QILLRIWFGLVWFGRVTLCCCSTKYKVLLN 565
>BI427218
Length = 421
Score = 34.3 bits (77), Expect = 0.14
Identities = 17/61 (27%), Positives = 30/61 (48%), Gaps = 6/61 (9%)
Frame = +1
Query: 305 LKARYYRRNAIFEANKGNNLSYVWKSIHEAIP------MVKQGTRWKIGNGDKIRVFQDR 358
L ++Y + E +GN S WK I+ P + +G +WK+ G+K + ++D
Sbjct: 55 LDSKYGDWRHLEEPTRGNRASAWWKDINIIDPSGADGSLFDKGIKWKVACGEKTKFWEDG 234
Query: 359 W 359
W
Sbjct: 235 W 237
>BM309783
Length = 433
Score = 32.7 bits (73), Expect = 0.40
Identities = 15/39 (38%), Positives = 22/39 (55%)
Frame = +3
Query: 322 NNLSYVWKSIHEAIPMVKQGTRWKIGNGDKIRVFQDRWL 360
N L +WKSI + P + RW + NG IR + D+W+
Sbjct: 231 NLLIPLWKSIVKL*PNIINVERWIVNNGRSIRAW*DKWV 347
>BI970631
Length = 775
Score = 32.0 bits (71), Expect = 0.68
Identities = 14/44 (31%), Positives = 21/44 (46%), Gaps = 5/44 (11%)
Frame = +1
Query: 325 SYVWKSI-----HEAIPMVKQGTRWKIGNGDKIRVFQDRWLVNG 363
S+ W+ + H ++ WK G GDKIR ++D W G
Sbjct: 493 SHWWRDLRKIYQHNDFKIIHXXMAWKAGCGDKIRFWKDNWXXXG 624
>BM891241
Length = 407
Score = 31.6 bits (70), Expect = 0.89
Identities = 18/60 (30%), Positives = 28/60 (46%), Gaps = 5/60 (8%)
Frame = -3
Query: 325 SYVWKSIHEAI-----PMVKQGTRWKIGNGDKIRVFQDRWLVNGGLAQPAA*YNRLLQLN 379
SY W+ + + + Q WK+G GDKI + D+WL + YN+L +N
Sbjct: 174 SYWWRDLRKFYHQSDHSIFHQYMSWKVGCGDKINFWTDKWLGEDYTLEQK--YNQLFLIN 1
>BI470652 similar to PIR|T17102|T171 RNA-directed DNA polymerase (EC
2.7.7.49) (clone AmLi2) - garden snapdragon (fragment),
partial (22%)
Length = 111
Score = 29.3 bits (64), Expect = 4.4
Identities = 12/34 (35%), Positives = 16/34 (46%)
Frame = +1
Query: 39 CVESVKYKVLVNQESEGPILPGSGLRKGDPLSPY 72
C+ S +LVN P GLR+G P P+
Sbjct: 7 CLSSASISILVNGSPTEEFKPXRGLRQGGPFGPF 108
>BM891637
Length = 427
Score = 29.3 bits (64), Expect = 4.4
Identities = 10/32 (31%), Positives = 19/32 (59%)
Frame = -1
Query: 332 HEAIPMVKQGTRWKIGNGDKIRVFQDRWLVNG 363
H+++ WK+G GD+I ++D W+ +G
Sbjct: 169 HQSMIAASNQFCWKVGRGDQILFWEDSWVDDG 74
>TC220428 weakly similar to UP|Q9FI28 (Q9FI28) Laccase (Diphenol oxidase),
partial (32%)
Length = 1045
Score = 28.9 bits (63), Expect = 5.8
Identities = 24/72 (33%), Positives = 35/72 (48%), Gaps = 6/72 (8%)
Frame = +3
Query: 303 DFLKARYYRRNAIFEANKGNNLSYVWKSIHEAIPMV----KQGTRWKIGN-GDKIR-VFQ 356
D LKA YY N ++E +++ + +P+ KQGTR + N G + VFQ
Sbjct: 303 DILKAYYYHINGVYEPGFPTFPPFIFNFTGDFLPITLNTPKQGTRVNVLNYGATVEIVFQ 482
Query: 357 DRWLVNGGLAQP 368
LV GG+ P
Sbjct: 483 GTNLV-GGIDHP 515
>TC226215 homologue to UP|Q6S4R9 (Q6S4R9) Allantoinase , partial (23%)
Length = 604
Score = 28.5 bits (62), Expect = 7.6
Identities = 16/50 (32%), Positives = 27/50 (54%)
Frame = +1
Query: 376 LQLNVKELLQPGQKSWNLDVLQHYLSSQSVFEAMNTLLFNSTAYDERVWR 425
LQL ++L G +SW+L + L S ++ +++LL Y E+ WR
Sbjct: 133 LQLETMQIL*FGNQSWSLTWMMIILFSSNI---LSSLLTWEEGYPEKFWR 273
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.344 0.154 0.514
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,987,711
Number of Sequences: 63676
Number of extensions: 320850
Number of successful extensions: 2982
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 2964
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2977
length of query: 520
length of database: 12,639,632
effective HSP length: 102
effective length of query: 418
effective length of database: 6,144,680
effective search space: 2568476240
effective search space used: 2568476240
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC147202.9


